The RBPome of influenza A virus NP-mRNA reveals a role for TDP-43 in viral replication
- PMID: 38686810
- PMCID: PMC11229366
- DOI: 10.1093/nar/gkae291
The RBPome of influenza A virus NP-mRNA reveals a role for TDP-43 in viral replication
Abstract
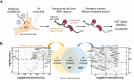
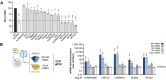
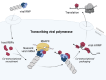
Genome-wide approaches have significantly advanced our knowledge of the repertoire of RNA-binding proteins (RBPs) that associate with cellular polyadenylated mRNAs within eukaryotic cells. Recent studies focusing on the RBP interactomes of viral mRNAs, notably SARS-Cov-2, have revealed both similarities and differences between the RBP profiles of viral and cellular mRNAs. However, the RBPome of influenza virus mRNAs remains unexplored. Herein, we identify RBPs that associate with the viral mRNA encoding the nucleoprotein (NP) of an influenza A virus. Focusing on TDP-43, we show that it binds several influenza mRNAs beyond the NP-mRNA, and that its depletion results in lower levels of viral mRNAs and proteins within infected cells, and a decreased yield of infectious viral particles. We provide evidence that the viral polymerase recruits TDP-43 onto viral mRNAs through a direct interaction with the disordered C-terminal domain of TDP-43. Notably, other RBPs found to be associated with influenza virus mRNAs also interact with the viral polymerase, which points to a role of the polymerase in orchestrating the assembly of viral messenger ribonucleoproteins.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

