Targeted deletion of Pf prophages from diverse Pseudomonas aeruginosa isolates has differential impacts on quorum sensing and virulence traits
- PMID: 38687034
- PMCID: PMC11112994
- DOI: 10.1128/jb.00402-23
Targeted deletion of Pf prophages from diverse Pseudomonas aeruginosa isolates has differential impacts on quorum sensing and virulence traits
Abstract
Pseudomonas aeruginosa is an opportunistic bacterial pathogen that commonly causes medical hardware, wound, and respiratory infections. Temperate filamentous Pf phages that infect P. aeruginosa impact numerous virulence phenotypes. Most work on Pf phages has focused on Pf4 and its host P. aeruginosa PAO1. Expanding from Pf4 and PAO1, this study explores diverse Pf phages infecting P. aeruginosa clinical isolates. We describe a simple technique targeting the Pf lysogeny maintenance gene, pflM (PA0718), that enables the effective elimination of Pf prophages from diverse P. aeruginosa hosts. The pflM gene shows diversity among different Pf phage isolates; however, all examined pflM alleles encode the DUF5447 domain. We demonstrate that pflM deletion results in prophage excision but not replication, leading to total prophage loss, indicating a role for lysis/lysogeny decisions for the DUF5447 domain. This study also assesses the effects different Pf phages have on host quorum sensing, biofilm formation, pigment production, and virulence against the bacterivorous nematode Caenorhabditis elegans. We find that Pf phages have strain-specific impacts on quorum sensing and biofilm formation, but nearly all suppress pigment production and increase C. elegans avoidance behavior. Collectively, this research not only introduces a valuable tool for Pf prophage elimination from diverse P. aeruginosa isolates but also advances our understanding of the complex relationship between P. aeruginosa and filamentous Pf phages.IMPORTANCEPseudomonas aeruginosa is an opportunistic bacterial pathogen that is frequently infected by filamentous Pf phages (viruses) that integrate into its chromosome, affecting behavior. Although prior work has focused on Pf4 and PAO1, this study investigates diverse Pf in clinical isolates. A simple method targeting the deletion of the Pf lysogeny maintenance gene pflM (PA0718) effectively eliminates Pf prophages from clinical isolates. The research evaluates the impact Pf prophages have on bacterial quorum sensing, biofilm formation, and virulence phenotypes. This work introduces a valuable tool to eliminate Pf prophages from clinical isolates and advances our understanding of P. aeruginosa and filamentous Pf phage interactions.
Keywords: C. elegans; Filamentous Pf bacteriophage; Pseudomonas aeruginosa; biofilms; prophage; pyocyanin; quorum sensing; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
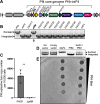
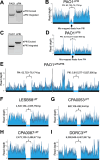

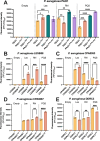
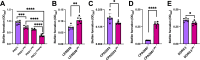
Update of
-
Targeted deletion of Pf prophages from diverse Pseudomonas aeruginosa isolates impacts quorum sensing and virulence traits.bioRxiv [Preprint]. 2023 Nov 19:2023.11.19.567716. doi: 10.1101/2023.11.19.567716. bioRxiv. 2023. Update in: J Bacteriol. 2024 May 23;206(5):e0040223. doi: 10.1128/jb.00402-23. PMID: 38014273 Free PMC article. Updated. Preprint.
Similar articles
-
Targeted deletion of Pf prophages from diverse Pseudomonas aeruginosa isolates impacts quorum sensing and virulence traits.bioRxiv [Preprint]. 2023 Nov 19:2023.11.19.567716. doi: 10.1101/2023.11.19.567716. bioRxiv. 2023. Update in: J Bacteriol. 2024 May 23;206(5):e0040223. doi: 10.1128/jb.00402-23. PMID: 38014273 Free PMC article. Updated. Preprint.
-
Phage Morons Play an Important Role in Pseudomonas aeruginosa Phenotypes.J Bacteriol. 2018 Oct 23;200(22):e00189-18. doi: 10.1128/JB.00189-18. Print 2018 Nov 15. J Bacteriol. 2018. PMID: 30150232 Free PMC article.
-
The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage.ISME J. 2009 Mar;3(3):271-82. doi: 10.1038/ismej.2008.109. Epub 2008 Nov 13. ISME J. 2009. PMID: 19005496 Free PMC article.
-
Phage against the Machine: The SIE-ence of Superinfection Exclusion.Viruses. 2024 Aug 23;16(9):1348. doi: 10.3390/v16091348. Viruses. 2024. PMID: 39339825 Free PMC article. Review.
-
Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections.Front Immunol. 2020 Feb 21;11:244. doi: 10.3389/fimmu.2020.00244. eCollection 2020. Front Immunol. 2020. PMID: 32153575 Free PMC article. Review.
Cited by
-
Polymicrobial interactions influence Mycobacterium abscessus co-existence and biofilm forming capabilities.Front Microbiol. 2024 Nov 25;15:1484510. doi: 10.3389/fmicb.2024.1484510. eCollection 2024. Front Microbiol. 2024. PMID: 39654682 Free PMC article.
-
New Pseudomonas infections drive Pf phage transmission in CF airways.JCI Insight. 2025 Apr 22;10(11):e188146. doi: 10.1172/jci.insight.188146. eCollection 2025 Jun 9. JCI Insight. 2025. PMID: 40261708 Free PMC article.
-
'Wild Type'.Microbiology (Reading). 2024 Aug;170(8):001495. doi: 10.1099/mic.0.001495. Microbiology (Reading). 2024. PMID: 39212644 Free PMC article. Review.
-
The lysogenic filamentous Pseudomonas bacteriophage phage Pf slows mucociliary transport.PNAS Nexus. 2024 Sep 9;3(9):pgae390. doi: 10.1093/pnasnexus/pgae390. eCollection 2024 Sep. PNAS Nexus. 2024. PMID: 39301510 Free PMC article.
-
Pseudomonas superinfection drives Pf phage transmission within airway infections in patients with cystic fibrosis.bioRxiv [Preprint]. 2025 Jan 14:2025.01.14.632786. doi: 10.1101/2025.01.14.632786. bioRxiv. 2025. PMID: 39868244 Free PMC article. Preprint.
References
-
- Secor PR, Burgener EB, Kinnersley M, Jennings LK, Roman-Cruz V, Popescu M, Van Belleghem JD, Haddock N, Copeland C, Michaels LA, de Vries CR, Chen Q, Pourtois J, Wheeler TJ, Milla CE, Bollyky PL. 2020. Pf Bacteriophage and their impact on Pseudomonas virulence, mammalian immunity, and chronic infections. Front Immunol 11:244. doi:10.3389/fimmu.2020.00244 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- R01AI138981/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P20 GM103546/GM/NIGMS NIH HHS/United States
- R01 HL148184/HL/NHLBI NIH HHS/United States
- 366502/NSF | National Science Foundation Graduate Research Fellowship Program (GRFP)
- P20GM103546/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources
Miscellaneous

