Novel multiplex-PCR test for Escherichia coli detection
- PMID: 38687052
- PMCID: PMC11237426
- DOI: 10.1128/spectrum.03773-23
Novel multiplex-PCR test for Escherichia coli detection
Abstract
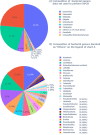
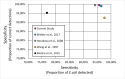
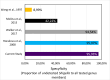
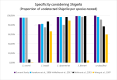
Escherichia coli is a diverse and ubiquitous strain of both commensal and pathogenic bacteria. In this study, we propose the use of multiplex polymerase chain reaction (PCR), using amplification of three genes (cydA, lacY, and ydiV), as a method for determining the affiliation of the tested strains to the E. coli species. The novelty of the method lies in the small number of steps needed to perform the diagnosis and, consequently, in the small amount of time needed to obtain it. This method, like any other, has some limitations, but its advantage is fast, cheap, and reliable identification of the presence of E. coli. Sequences of the indicated genes from 1,171 complete E. coli genomes in the NCBI database were used to prepare the primers. The developed multiplex PCR was tested on 47,370 different Enterobacteriaceae genomes using in silico PCR. The sensitivity and specificity of the developed test were 95.76% and 99.49%, respectively. Wet laboratory analyses confirmed the high specificity, repeatability, reproducibility, and reliability of the proposed test. Because of the detection of three genes, this method is very cost and labor-effective, yet still highly accurate, specific, and sensitive in comparison to similar methods.
Importance: Detection of E. coli from environmental or clinical samples is important due to the common occurrence of this species of bacteria in all human and animal environments. As commonly known, these bacteria strains can be commensal and pathogenic, causing numerous infections of clinical importance, including infections of the digestive system, urinary, respiratory, and even meninges, particularly dangerous for newborns. The developed multiplex polymerase chain reaction test, confirming the presence of E. coli in samples, can be used in many laboratories. The test provides new opportunities for quick and cheap analyses, detecting E. coli using only three pairs of primers (analysis of the presence of three genes) responsible for metabolism and distinguishing E. coli from other pathogens from the Enterobacteriaceae family. Compared to other tests previously described in the literature, our method is characterized by high specificity and sensitivity.
Keywords: Escherichia coli; PCR; detection; diagnostics; multiplex PCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Hacker J, Blum-Oehler G. 2007. In appreciation of Theodor Escherich. Nat Rev Microbiol 5:902–902. doi: 10.1038/nrmicro1810 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

