Androgen receptor splice variants drive castration-resistant prostate cancer metastasis by activating distinct transcriptional programs
- PMID: 38687617
- PMCID: PMC11142739
- DOI: 10.1172/JCI168649
Androgen receptor splice variants drive castration-resistant prostate cancer metastasis by activating distinct transcriptional programs
Abstract
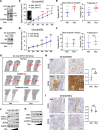
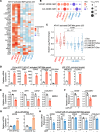
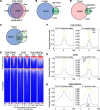
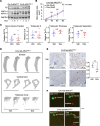
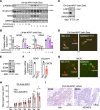
One critical mechanism through which prostate cancer (PCa) adapts to treatments targeting androgen receptor (AR) signaling is the emergence of ligand-binding domain-truncated and constitutively active AR splice variants, particularly AR-V7. While AR-V7 has been intensively studied, its ability to activate distinct biological functions compared with the full-length AR (AR-FL), and its role in regulating the metastatic progression of castration-resistant PCa (CRPC), remain unclear. Our study found that, under castrated conditions, AR-V7 strongly induced osteoblastic bone lesions, a response not observed with AR-FL overexpression. Through combined ChIP-seq, ATAC-seq, and RNA-seq analyses, we demonstrated that AR-V7 uniquely accesses the androgen-responsive elements in compact chromatin regions, activating a distinct transcription program. This program was highly enriched for genes involved in epithelial-mesenchymal transition and metastasis. Notably, we discovered that SOX9, a critical metastasis driver gene, was a direct target and downstream effector of AR-V7. Its protein expression was dramatically upregulated in AR-V7-induced bone lesions. Moreover, we found that Ser81 phosphorylation enhanced AR-V7's pro-metastasis function by selectively altering its specific transcription program. Blocking this phosphorylation with CDK9 inhibitors impaired the AR-V7-mediated metastasis program. Overall, our study has provided molecular insights into the role of AR splice variants in driving the metastatic progression of CRPC.
Keywords: Molecular genetics; Oncology; Prostate cancer; Transcription.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

