ER-associated degradation adapter Sel1L is required for CD8+ T cell function and memory formation following acute viral infection
- PMID: 38687642
- PMCID: PMC11194752
- DOI: 10.1016/j.celrep.2024.114156
ER-associated degradation adapter Sel1L is required for CD8+ T cell function and memory formation following acute viral infection
Abstract
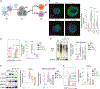
The maintenance of antigen-specific CD8+ T cells underlies the efficacy of vaccines and immunotherapies. Pathways contributing to CD8+ T cell loss are not completely understood. Uncovering the pathways underlying the limited persistence of CD8+ T cells would be of significant benefit for developing novel strategies of promoting T cell persistence. Here, we demonstrate that murine CD8+ T cells experience endoplasmic reticulum (ER) stress following activation and that the ER-associated degradation (ERAD) adapter Sel1L is induced in activated CD8+ T cells. Sel1L loss limits CD8+ T cell function and memory formation following acute viral infection. Mechanistically, Sel1L is required for optimal bioenergetics and c-Myc expression. Finally, we demonstrate that human CD8+ T cells experience ER stress upon activation and that ER stress is negatively associated with improved T cell functionality in T cell-redirecting therapies. Together, these results demonstrate that ER stress and ERAD are important regulators of T cell function and persistence.
Keywords: CD8 T cell; CP: Immunology; ER stress; ERAD; Myc; T cell memory; immunometabolism; protein homeostasis.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests In the past three years, C.A.L. has consulted for Astellas Pharmaceuticals, Odyssey Therapeutics, Third Rock Ventures, and T-Knife Therapeutics and is an inventor on patents pertaining to Kras-regulated metabolic pathways, redox control pathways in pancreatic cancer, and therapeutically targeting the GOT1-ME1 pathway (US Patent no. 2015126580-A1, 05/07/2015; US Patent no. 20190136238, 05/09/2019; International Patent no. WO2013177426-A2, 04/23/2015).
Figures




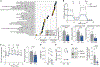

References
Publication types
MeSH terms
Substances
Grants and funding
- F99 AG079793/AG/NIA NIH HHS/United States
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- 75N93022D00005/AI/NIAID NIH HHS/United States
- R01 AI165533/AI/NIAID NIH HHS/United States
- 75N95020D00005/DA/NIDA NIH HHS/United States
- T32 AI007413/AI/NIAID NIH HHS/United States
- T32 CA140044/CA/NCI NIH HHS/United States
- R01 AI157384/AI/NIAID NIH HHS/United States
- 75N93023D00005/AI/NIAID NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- F31 NS124280/NS/NINDS NIH HHS/United States
- 75N99020D00005/OF/ORFDO NIH HHS/United States
- R01 NS120322/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

