Genomic deletion of Bcl6 differentially affects conventional dendritic cell subsets and compromises Tfh/Tfr/Th17 cell responses
- PMID: 38688934
- PMCID: PMC11061177
- DOI: 10.1038/s41467-024-46966-6
Genomic deletion of Bcl6 differentially affects conventional dendritic cell subsets and compromises Tfh/Tfr/Th17 cell responses
Abstract
Conventional dendritic cells (cDC) play key roles in immune induction, but what drives their heterogeneity and functional specialization is still ill-defined. Here we show that cDC-specific deletion of the transcriptional repressor Bcl6 in mice alters the phenotype and transcriptome of cDC1 and cDC2, while their lineage identity is preserved. Bcl6-deficient cDC1 are diminished in the periphery but maintain their ability to cross-present antigen to CD8+ T cells, confirming general maintenance of this subset. Surprisingly, the absence of Bcl6 in cDC causes a complete loss of Notch2-dependent cDC2 in the spleen and intestinal lamina propria. DC-targeted Bcl6-deficient mice induced fewer T follicular helper cells despite a profound impact on T follicular regulatory cells in response to immunization and mounted diminished Th17 immunity to Citrobacter rodentium in the colon. Our findings establish Bcl6 as an essential transcription factor for subsets of cDC and add to our understanding of the transcriptional landscape underlying cDC heterogeneity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






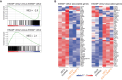
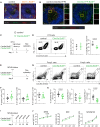


References
Publication types
MeSH terms
Substances
Grants and funding
- 2020-01977/Vetenskapsrådet (Swedish Research Council)
- R215-2015-4100/Lundbeckfonden (Lundbeck Foundation)
- 21 1826 Pj/Cancerfonden (Swedish Cancer Society)
- 252623821/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Schr 1444/1-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 360372040/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 322359157-FOR2599-A03/Deutsche Forschungsgemeinschaft (German Research Foundation)
- BA 5132/1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 210592381/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 390873048/Deutsche Forschungsgemeinschaft (German Research Foundation)
- ERC-2016-STG-715182/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- ERC-2016-STG-715182/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

