Targeting pyruvate dehydrogenase kinase 1 overcomes EGFR C797S mutation-driven osimertinib resistance in non-small cell lung cancer
- PMID: 38689087
- PMCID: PMC11148081
- DOI: 10.1038/s12276-024-01221-2
Targeting pyruvate dehydrogenase kinase 1 overcomes EGFR C797S mutation-driven osimertinib resistance in non-small cell lung cancer
Abstract
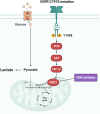
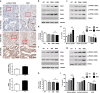
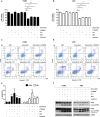
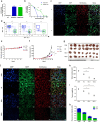
Osimertinib, a selective third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI), effectively targets the EGFR T790M mutant in non-small cell lung cancer (NSCLC). However, the newly identified EGFR C797S mutation confers resistance to osimertinib. In this study, we explored the role of pyruvate dehydrogenase kinase 1 (PDK1) in osimertinib resistance. Patients exhibiting osimertinib resistance initially displayed elevated PDK1 expression. Osimertinib-resistant cell lines with the EGFR C797S mutation were established using A549, NCI-H292, PC-9, and NCI-H1975 NSCLC cells for both in vitro and in vivo investigations. These EGFR C797S mutant cells exhibited heightened phosphorylation of EGFR, leading to the activation of downstream oncogenic pathways. The EGFR C797S mutation appeared to increase PDK1-driven glycolysis through the EGFR/AKT/HIF-1α axis. Combining osimertinib with the PDK1 inhibitor leelamine helped successfully overcome osimertinib resistance in allograft models. CRISPR-mediated PDK1 knockout effectively inhibited tumor formation in xenograft models. Our study established a clear link between the EGFR C797S mutation and elevated PDK1 expression, opening new avenues for the discovery of targeted therapies and improving our understanding of the roles of EGFR mutations in cancer progression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

