Unveiling microbial diversity: harnessing long-read sequencing technology
- PMID: 38689099
- PMCID: PMC11955098
- DOI: 10.1038/s41592-024-02262-1
Unveiling microbial diversity: harnessing long-read sequencing technology
Abstract
Long-read sequencing has recently transformed metagenomics, enhancing strain-level pathogen characterization, enabling accurate and complete metagenome-assembled genomes, and improving microbiome taxonomic classification and profiling. These advancements are not only due to improvements in sequencing accuracy, but also happening across rapidly changing analysis methods. In this Review, we explore long-read sequencing's profound impact on metagenomics, focusing on computational pipelines for genome assembly, taxonomic characterization and variant detection, to summarize recent advancements in the field and provide an overview of available analytical methods to fully leverage long reads. We provide insights into the advantages and disadvantages of long reads over short reads and their evolution from the early days of long-read sequencing to their recent impact on metagenomics and clinical diagnostics. We further point out remaining challenges for the field such as the integration of methylation signals in sub-strain analysis and the lack of benchmarks.
© 2024. Springer Nature America, Inc.
Conflict of interest statement
Competing interests
F.J.S. has received research funding from Illumina, PacBio, Genentech and Oxford Nanopore. V.K.M. is an employee of Genentech. The remaining authors declare no competing interests.
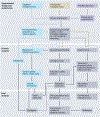
Figures


References
-
- Sedlazeck FJ, Lee H, Darby CA & Schatz MC Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 19, 329–346 (2018). - PubMed
Publication types
MeSH terms
Grants and funding
- P01 AI152999/AI/NIAID NIH HHS/United States
- 1U19AI144297/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- EF-2126387/National Science Foundation (NSF)
- III-2239114/National Science Foundation (NSF)
- U19 AI144297/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

