OGNNMDA: a computational model for microbe-drug association prediction based on ordered message-passing graph neural networks
- PMID: 38689654
- PMCID: PMC11058190
- DOI: 10.3389/fgene.2024.1370013
OGNNMDA: a computational model for microbe-drug association prediction based on ordered message-passing graph neural networks
Abstract
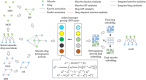
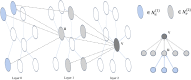
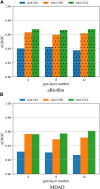
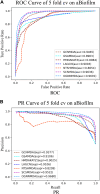
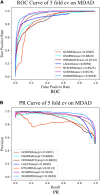
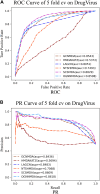
In recent years, many excellent computational models have emerged in microbe-drug association prediction, but their performance still has room for improvement. This paper proposed the OGNNMDA framework, which applied an ordered message-passing mechanism to distinguish the different neighbor information in each message propagation layer, and it achieved a better embedding ability through deeper network layers. Firstly, the method calculates four similarity matrices based on microbe functional similarity, drug chemical structure similarity, and their respective Gaussian interaction profile kernel similarity. After integrating these similarity matrices, it concatenates the integrated similarity matrix with the known association matrix to obtain the microbe-drug heterogeneous matrix. Secondly, it uses a multi-layer ordered message-passing graph neural network encoder to encode the heterogeneous network and the known association information adjacency matrix, thereby obtaining the final embedding features of the microbe-drugs. Finally, it inputs the embedding features into the bilinear decoder to get the final prediction results. The OGNNMDA method performed comparative experiments, ablation experiments, and case studies on the aBiofilm, MDAD and DrugVirus datasets using 5-fold cross-validation. The experimental results showed that OGNNMDA showed the strongest prediction performance on aBiofilm and MDAD and obtained sub-optimal results on DrugVirus. In addition, the case studies on well-known drugs and microbes also support the effectiveness of the OGNNMDA method. Source codes and data are available at: https://github.com/yyzg/OGNNMDA.
Keywords: graph neural network; microbe-drug association; multi-similarities; ordered message-passing mechanism; prediction model.
Copyright © 2024 Zhao, Kuang, Hu, Zhang, Yang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Prediction of Human Microbe-Drug Association based on Layer Attention Graph Convolutional Network.Curr Med Chem. 2024;31(31):5097-5109. doi: 10.2174/0109298673249941231108091326. Curr Med Chem. 2024. PMID: 39225188
-
Neighborhood-based inference and restricted Boltzmann machine for microbe and drug associations prediction.PeerJ. 2022 Aug 15;10:e13848. doi: 10.7717/peerj.13848. eCollection 2022. PeerJ. 2022. PMID: 35990901 Free PMC article.
-
HKFGCN: A novel multiple kernel fusion framework on graph convolutional network to predict microbe-drug associations.Comput Biol Chem. 2024 Jun;110:108041. doi: 10.1016/j.compbiolchem.2024.108041. Epub 2024 Mar 2. Comput Biol Chem. 2024. PMID: 38471354
-
GLNNMDA: a multimodal prediction model for microbe-drug associations based on global and local features.Sci Rep. 2024 Sep 6;14(1):20847. doi: 10.1038/s41598-024-71837-x. Sci Rep. 2024. PMID: 39242712 Free PMC article.
-
DMGL-MDA: A dual-modal graph learning method for microbe-drug association prediction.Methods. 2024 Feb;222:51-56. doi: 10.1016/j.ymeth.2023.12.005. Epub 2024 Jan 4. Methods. 2024. PMID: 38184219
Cited by
-
Prediction of microbe-drug associations using a CNN-Bernoulli random forest model.PeerJ. 2025 Aug 5;13:e19637. doi: 10.7717/peerj.19637. eCollection 2025. PeerJ. 2025. PMID: 40786104 Free PMC article.
-
A deep drug prediction framework for viral infectious diseases using an optimizer-based ensemble of convolutional neural network: COVID-19 as a case study.Mol Divers. 2025 Jun;29(3):2473-2487. doi: 10.1007/s11030-024-11003-7. Epub 2024 Oct 9. Mol Divers. 2025. PMID: 39379663
References
-
- Berg R. V. D., Kipf T. N., Welling M. (2017). Graph convolutional matrix completion. arXiv preprint arXiv:1706.02263.
LinkOut - more resources
Full Text Sources

