Comparative analysis of 10X Chromium vs. BD Rhapsody whole transcriptome single-cell sequencing technologies in complex human tissues
- PMID: 38689972
- PMCID: PMC11059509
- DOI: 10.1016/j.heliyon.2024.e28358
Comparative analysis of 10X Chromium vs. BD Rhapsody whole transcriptome single-cell sequencing technologies in complex human tissues
Abstract
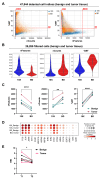
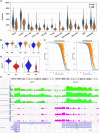
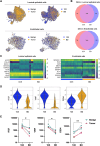
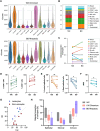
The development of single-cell omics tools has enabled scientists to study the tumor microenvironment (TME) in unprecedented detail. However, each of the different techniques may have its unique strengths and limitations. Here we directly compared two commercially available high-throughput single-cell RNA sequencing (scRNA-seq) technologies - droplet-based 10X Chromium vs. microwell-based BD Rhapsody - using paired samples from patients with localized prostate cancer (PCa) undergoing a radical prostatectomy. Although high technical consistency was observed in unraveling the whole transcriptome, the relative abundance of cell populations differed. Cells with low mRNA content such as T cells were underrepresented in the droplet-based system, at least partly due to lower RNA capture rates. In contrast, microwell-based scRNA-seq recovered less cells of epithelial origin. Moreover, we discovered platform-dependent variabilities in mRNA quantification and cell-type marker annotation. Overall, our study provides important information for selection of the appropriate scRNA-seq platform and for the interpretation of published results.
Keywords: 10X Chromium; BD Rhapsody; Low-mRNA content cells; Neutrophils; Prostate cancer; Single-cell RNA sequencing.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nature medicine. 2018;24:1277–1289. - PubMed
-
- Guo X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nature medicine. 2018;24:978–985. - PubMed
-
- Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845 e820. - PubMed
-
- Goveia J., Rohlenova K., Taverna F., Treps L., Conradi L.C., Pircher A., et al. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell. 2020;37:421. - PubMed
LinkOut - more resources
Full Text Sources

