Role of proteoglycan synthesis genes in osteosarcoma stem cells
- PMID: 38690160
- PMCID: PMC11058990
- DOI: 10.3389/fonc.2024.1325794
Role of proteoglycan synthesis genes in osteosarcoma stem cells
Abstract
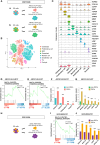
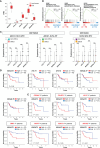
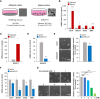
Osteosarcoma stem cells (OSCs) contribute to the pathogenesis of osteosarcoma (OS), which is the most common malignant primary bone tumor. The significance and underlying mechanisms of action of proteoglycans (PGs) and glycosaminoglycans (GAGs) in OSC phenotypes and OS malignancy are largely unknown. This study aimed to investigate the role of PG/GAG biosynthesis and the corresponding candidate genes in OSCs and poor clinical outcomes in OS using scRNA-seq and bulk RNA-seq datasets of clinical OS specimens, accompanied by biological validation by in vitro genetic and pharmacological analyses. The expression of β-1,3-glucuronyltransferase 3 (B3GAT3), one of the genes responsible for the biosynthesis of the common core tetrasaccharide linker region of PGs, was significantly upregulated in both OSC populations and OS tissues and was associated with poor survival in patients with OS with high stem cell properties. Moreover, the genetic inactivation of B3GAT3 by RNA interference and pharmacological inhibition of PG biosynthesis abrogated the self-renewal potential of OSCs. Collectively, these findings suggest a pivotal role for B3GAT3 and PG/GAG biosynthesis in the regulation of OSC phenotypes and OS malignancy, thereby providing a potential target for OSC-directed therapy.
Keywords: 3glucuronyltransferase 3; glycosaminoglycan; osteosarcoma; osteosarcoma stem cell; proteoglycan; β-1.
Copyright © 2024 Osumi, Sugihara, Yoshimoto, Tokumura, Tanaka and Hinoi.
Conflict of interest statement
EH received grants from the Japan Society for the Promotion of Science for this study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials

