The natural history and genotype-phenotype correlations of TMPRSS3 hearing loss: an international, multi-center, cohort analysis
- PMID: 38691166
- PMCID: PMC11098735
- DOI: 10.1007/s00439-024-02648-3
The natural history and genotype-phenotype correlations of TMPRSS3 hearing loss: an international, multi-center, cohort analysis
Abstract
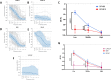
TMPRSS3-related hearing loss presents challenges in correlating genotypic variants with clinical phenotypes due to the small sample sizes of previous studies. We conducted a cross-sectional genomics study coupled with retrospective clinical phenotype analysis on 127 individuals. These individuals were from 16 academic medical centers across 6 countries. Key findings revealed 47 unique TMPRSS3 variants with significant differences in hearing thresholds between those with missense variants versus those with loss-of-function genotypes. The hearing loss progression rate for the DFNB8 subtype was 0.3 dB/year. Post-cochlear implantation, an average word recognition score of 76% was observed. Of the 51 individuals with two missense variants, 10 had DFNB10 with profound hearing loss. These 10 all had at least one of 4 TMPRSS3 variants predicted by computational modeling to be damaging to TMPRSS3 structure and function. To our knowledge, this is the largest study of TMPRSS3 genotype-phenotype correlations. We find significant differences in hearing thresholds, hearing loss progression, and age of presentation, by TMPRSS3 genotype and protein domain affected. Most individuals with TMPRSS3 variants perform well on speech recognition tests after cochlear implant, however increased age at implant is associated with worse outcomes. These findings provide insight for genetic counseling and the on-going design of novel therapeutic approaches.
© 2024. The Author(s).
Conflict of interest statement
Xue Z. Liu is an advisor to Salubritas Therapeutics. Xue Z. Liu is founding advisor and Jeffery Holt is a Scientific advisor to Rescue Hearing Inc., which is involved in developing
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

