PRMT1 promotes epigenetic reprogramming associated with acquired chemoresistance in pancreatic cancer
- PMID: 38691454
- PMCID: PMC11238875
- DOI: 10.1016/j.celrep.2024.114176
PRMT1 promotes epigenetic reprogramming associated with acquired chemoresistance in pancreatic cancer
Abstract
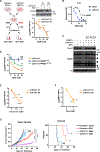
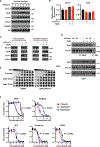
Pancreatic ductal adenocarcinoma (PDAC) carries a dismal prognosis due to therapeutic resistance. We show that PDAC cells undergo global epigenetic reprogramming to acquire chemoresistance, a process that is driven at least in part by protein arginine methyltransferase 1 (PRMT1). Genetic or pharmacological PRMT1 inhibition impairs adaptive epigenetic reprogramming and delays acquired resistance to gemcitabine and other common chemo drugs. Mechanistically, gemcitabine treatment induces translocation of PRMT1 into the nucleus, where its enzymatic activity limits the assembly of chromatin-bound MAFF/BACH1 transcriptional complexes. Cut&Tag chromatin profiling of H3K27Ac, MAFF, and BACH1 suggests a pivotal role for MAFF/BACH1 in global epigenetic response to gemcitabine, which is confirmed by genetically silencing MAFF. PRMT1 and MAFF/BACH1 signature genes identified by Cut&Tag analysis distinguish gemcitabine-resistant from gemcitabine-sensitive patient-derived xenografts of PDAC, supporting the PRMT1-MAFF/BACH1 epigenetic regulatory axis as a potential therapeutic avenue for improving the efficacy and durability of chemotherapies in patients of PDAC.
Keywords: BACH1; CP: Cancer; MAFF; PRMT1; chemoresistance; epigenetic reprogramming; pancreatic cancer.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Similar articles
-
PRMT1 promotes pancreatic cancer development and resistance to chemotherapy.Cell Rep Med. 2024 Mar 19;5(3):101461. doi: 10.1016/j.xcrm.2024.101461. Epub 2024 Mar 8. Cell Rep Med. 2024. PMID: 38460517 Free PMC article.
-
PRMT1 promotes pancreatic cancer growth and predicts poor prognosis.Cell Oncol (Dordr). 2020 Feb;43(1):51-62. doi: 10.1007/s13402-019-00435-1. Epub 2019 Sep 13. Cell Oncol (Dordr). 2020. PMID: 31520395
-
The interaction between UBR7 and PRMT5 drives PDAC resistance to gemcitabine by regulating glycolysis and immune microenvironment.Cell Death Dis. 2024 Oct 18;15(10):758. doi: 10.1038/s41419-024-07145-z. Cell Death Dis. 2024. PMID: 39424627 Free PMC article.
-
Gemcitabine resistance in pancreatic ductal adenocarcinoma.Drug Resist Updat. 2015 Nov;23:55-68. doi: 10.1016/j.drup.2015.10.002. Epub 2015 Nov 3. Drug Resist Updat. 2015. PMID: 26690340 Review.
-
Protein Arginine Methyltransferase 1: A Multi-Purpose Player in the Development of Cancer and Metabolic Disease.Biomolecules. 2025 Jan 27;15(2):185. doi: 10.3390/biom15020185. Biomolecules. 2025. PMID: 40001488 Free PMC article. Review.
Cited by
-
Enhancer reprogramming: critical roles in cancer and promising therapeutic strategies.Cell Death Discov. 2025 Mar 3;11(1):84. doi: 10.1038/s41420-025-02366-3. Cell Death Discov. 2025. PMID: 40032852 Free PMC article. Review.
-
The Multifaceted Roles of BACH1 in Disease: Implications for Biological Functions and Therapeutic Applications.Adv Sci (Weinh). 2025 Mar;12(10):e2412850. doi: 10.1002/advs.202412850. Epub 2025 Jan 30. Adv Sci (Weinh). 2025. PMID: 39887888 Free PMC article. Review.
-
MAFF alleviates hepatic ischemia-reperfusion injury by regulating the CLCF1/STAT3 signaling pathway.Cell Mol Biol Lett. 2025 Apr 1;30(1):39. doi: 10.1186/s11658-025-00721-x. Cell Mol Biol Lett. 2025. PMID: 40169936 Free PMC article.
-
PRMT1 oligomerization regulates RNA-binding protein cascade to promote pancreatic cancer.Life Sci Alliance. 2025 Jul 17;8(9):e202503202. doi: 10.26508/lsa.202503202. Print 2025 Sep. Life Sci Alliance. 2025. PMID: 40675804 Free PMC article.
-
Phosphorylation of USP32 by CDK5 regulates Rap1 stability and therapeutic resistance in pancreatic ductal adenocarcinoma.Oncogene. 2025 Aug;44(30):2634-2645. doi: 10.1038/s41388-024-03263-2. Epub 2025 May 16. Oncogene. 2025. PMID: 40379759
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

