Pediatric Chordoma: A Tale of Two Genomes
- PMID: 38691518
- PMCID: PMC11296893
- DOI: 10.1158/1541-7786.MCR-23-0741
Pediatric Chordoma: A Tale of Two Genomes
Abstract
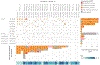
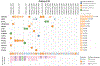
Little is known about the genomic alterations in chordoma, with the exception of loss of SMARCB1, a core member of the SWI/SNF complex, in poorly differentiated chordomas. A TBXT duplication and rs2305089 polymorphism, located at 6q27, are known genetic susceptibility loci. A comprehensive genomic analysis of the nuclear and mitochondrial genomes in pediatric chordoma has not yet been reported. In this study, we performed WES and mtDNA genome sequencing on 29 chordomas from 23 pediatric patients. Findings were compared with that from whole-genome sequencing datasets of 80 adult patients with skull base chordoma. In the pediatric chordoma cohort, 81% of the somatic mtDNA mutations were observed in NADH complex genes, which is significantly enriched compared with the rest of the mtDNA genes (P = 0.001). In adult chordomas, mtDNA mutations were also enriched in the NADH complex genes (P < 0.0001). Furthermore, a progressive increase in heteroplasmy of nonsynonymous mtDNA mutations was noted in patients with multiple tumors (P = 0.0007). In the nuclear genome, rare likely germline in-frame indels in ARID1B, a member of the SWI/SNF complex located at 6q25.3, were observed in five pediatric patients (22%) and four patients in the adult cohort (5%). The frequency of rare ARID1B indels in the pediatric cohort is significantly higher than that in the adult cohort (P = 0.0236, Fisher's exact test), but they were both significantly higher than that in the ethnicity-matched populations (P < 5.9e-07 and P < 0.0001174, respectively). Implications: germline ARID1B indels and mtDNA aberrations seem important for chordoma genesis, especially in pediatric chordoma.
©2024 American Association for Cancer Research.
Conflict of interest statement
Figures



References
-
- Lau CS, et al., Pediatric Chordomas: A Population-Based Clinical Outcome Study Involving 86 Patients from the Surveillance, Epidemiology, and End Result (SEER) Database (1973–2011). Pediatr Neurosurg, 2016. 51(3): p. 127–36. - PubMed
-
- Zhou J, et al., Prognostic Factors in Patients With Spinal Chordoma: An Integrative Analysis of 682 Patients. Neurosurgery, 2017. 81(5): p. 812–823. - PubMed
-
- Mobley BC, et al., Loss of SMARCB1/INI1 expression in poorly differentiated chordomas. Acta Neuropathol, 2010. 120(6): p. 745–53. - PubMed
-
- Hasselblatt M, et al., Poorly differentiated chordoma with SMARCB1/INI1 loss: a distinct molecular entity with dismal prognosis. Acta Neuropathol, 2016. 132(1): p. 149–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

