Exchange of subtelomeric regions between chromosomes 4q and 10q reverts the FSHD genotype and phenotype
- PMID: 38691604
- PMCID: PMC11062572
- DOI: 10.1126/sciadv.adl1922
Exchange of subtelomeric regions between chromosomes 4q and 10q reverts the FSHD genotype and phenotype
Abstract
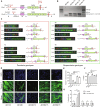
The most common form of facioscapulohumeral dystrophy (FSHD1) is caused by a partial loss of the D4Z4 macrosatellite repeat array in the subtelomeric region of chromosome 4. Patients with FSHD1 typically carry 1 to 10 D4Z4 repeats, whereas nonaffected individuals have 11 to 150 repeats. The ~150-kilobyte subtelomeric region of the chromosome 10q exhibits a ~99% sequence identity to the 4q, including the D4Z4 array. Nevertheless, contractions of the chr10 array do not cause FSHD or any known disease, as in most people D4Z4 array on chr10 is flanked by the nonfunctional polyadenylation signal, not permitting the DUX4 expression. Here, we attempted to correct the FSHD genotype by a CRISPR-Cas9-induced exchange of the chr4 and chr10 subtelomeric regions. We demonstrated that the induced t(4;10) translocation can generate recombinant genotypes translated into improved FSHD phenotype. FSHD myoblasts with the t(4;10) exhibited reduced expression of the DUX4 targets, restored PAX7 target expression, reduced sensitivity to oxidative stress, and improved differentiation capacity.
Figures


References
-
- Geng L. N., Yao Z., Snider L., Fong A. P., Cech J. N., Young J. M., van der Maarel S. M., Ruzzo W. L., Gentleman R. C., Tawil R., Tapscott S. J., DUX4 activates germline genes, retroelements, and immune mediators: Implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51 (2012). - PMC - PubMed
-
- Bosnakovski D., Xu Z., Gang E. J., Galindo C. L., Liu M., Simsek T., Garner H. R., Agha-Mohammadi S., Tassin A., Coppée F., Belayew A., Perlingeiro R. R., Kyba M., An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 27, 2766–2779 (2008). - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

