SETDB1 modulates the TGFβ response in Duchenne muscular dystrophy myotubes
- PMID: 38691608
- PMCID: PMC11062573
- DOI: 10.1126/sciadv.adj8042
SETDB1 modulates the TGFβ response in Duchenne muscular dystrophy myotubes
Abstract
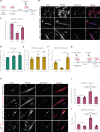
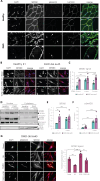
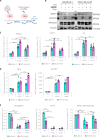
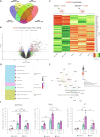
Overactivation of the transforming growth factor-β (TGFβ) signaling in Duchenne muscular dystrophy (DMD) is a major hallmark of disease progression, leading to fibrosis and muscle dysfunction. Here, we investigated the role of SETDB1 (SET domain, bifurcated 1), a histone lysine methyltransferase involved in muscle differentiation. Our data show that, following TGFβ induction, SETDB1 accumulates in the nuclei of healthy myotubes while being already present in the nuclei of DMD myotubes where TGFβ signaling is constitutively activated. Transcriptomics revealed that depletion of SETDB1 in DMD myotubes leads to down-regulation of TGFβ target genes coding for secreted factors involved in extracellular matrix remodeling and inflammation. Consequently, SETDB1 silencing in DMD myotubes abrogates the deleterious effect of their secretome on myoblast differentiation by impairing myoblast pro-fibrotic response. Our findings indicate that SETDB1 potentiates the TGFβ-driven fibrotic response in DMD muscles, providing an additional axis for therapeutic intervention.
Figures
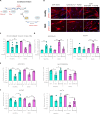
References
-
- Mozzetta C., Boyarchuk E., Pontis J., Ait-Si-Ali S., Sound of silence: The properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 16, 499–513 (2015). - PubMed
-
- Juznic L., Peuker K., Strigli A., Brosch M., Herrmann A., Hasler R., Koch M., Matthiesen L., Zeissig Y., Loscher B. S., Nuber A., Schotta G., Neumeister V., Chavakis T., Kurth T., Lesche M., Dahl A., von Massenhausen A., Linkermann A., Schreiber S., Aden K., Rosenstiel P. C., Franke A., Hampe J., Zeissig S., SETDB1 is required for intestinal epithelial differentiation and the prevention of intestinal inflammation. Gut 70, 485–498 (2021). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

