Vaccination with antigenically complex hemagglutinin mixtures confers broad protection from influenza disease
- PMID: 38691617
- PMCID: PMC12333966
- DOI: 10.1126/scitranslmed.adj4685
Vaccination with antigenically complex hemagglutinin mixtures confers broad protection from influenza disease
Abstract
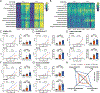
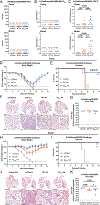
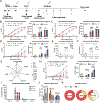
Current seasonal influenza virus vaccines induce responses primarily against immunodominant but highly plastic epitopes in the globular head of the hemagglutinin (HA) glycoprotein. Because of viral antigenic drift at these sites, vaccines need to be updated and readministered annually. To increase the breadth of influenza vaccine-mediated protection, we developed an antigenically complex mixture of recombinant HAs designed to redirect immune responses to more conserved domains of the protein. Vaccine-induced antibodies were disproportionally redistributed to the more conserved stalk of the HA without hindering, and in some cases improving, antibody responses against the head domain. These improved responses led to increased protection against homologous and heterologous viral challenges in both mice and ferrets compared with conventional vaccine approaches. Thus, antigenically complex protein mixtures can at least partially overcome HA head domain antigenic immunodominance and may represent a step toward a more universal influenza vaccine.
Conflict of interest statement
Figures



References
-
- World Health Organization, Influenza (Seasonal) (World Health Organization 2023); https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal).
-
- Black S, Nicolay U, Vesikari T, Knuf M, Del Giudice G, Della Cioppa G, Tsai T, Clemens R, Rappuoli R, Hemagglutination inhibition antibody titers as a correlate of protection for inactivated influenza vaccines in children. Pediatr Infect Dis J 30, 1081–1085 (2011); published online EpubDec ( 10.1097/INF.0b013e3182367662). - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

