Tumor mitochondrial oxidative phosphorylation stimulated by the nuclear receptor RORγ represents an effective therapeutic opportunity in osteosarcoma
- PMID: 38692271
- PMCID: PMC11148566
- DOI: 10.1016/j.xcrm.2024.101519
Tumor mitochondrial oxidative phosphorylation stimulated by the nuclear receptor RORγ represents an effective therapeutic opportunity in osteosarcoma
Abstract
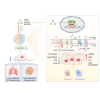
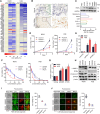
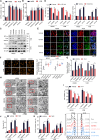
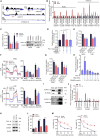
Osteosarcoma (OS) is the most common malignant bone tumor with a poor prognosis. Here, we show that the nuclear receptor RORγ may serve as a potential therapeutic target in OS. OS exhibits a hyperactivated oxidative phosphorylation (OXPHOS) program, which fuels the carbon source to promote tumor progression. We found that RORγ is overexpressed in OS tumors and is linked to hyperactivated OXPHOS. RORγ induces the expression of PGC-1β and physically interacts with it to activate the OXPHOS program by upregulating the expression of respiratory chain component genes. Inhibition of RORγ strongly inhibits OXPHOS activation, downregulates mitochondrial functions, and increases ROS production, which results in OS cell apoptosis and ferroptosis. RORγ inverse agonists strongly suppressed OS tumor growth and progression and sensitized OS tumors to chemotherapy. Taken together, our results indicate that RORγ is a critical regulator of the OXPHOS program in OS and provides an effective therapeutic strategy for this deadly disease.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

