Nanopore sequencing of influenza A and B in Oxfordshire and the United Kingdom, 2022-23
- PMID: 38692359
- PMCID: PMC11101610
- DOI: 10.1016/j.jinf.2024.106164
Nanopore sequencing of influenza A and B in Oxfordshire and the United Kingdom, 2022-23
Abstract
Objectives: We evaluated Nanopore sequencing for influenza surveillance.
Methods: Influenza A and B PCR-positive samples from hospital patients in Oxfordshire, UK, and a UK-wide population survey from winter 2022-23 underwent Nanopore sequencing following targeted rt-PCR amplification.
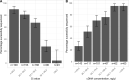
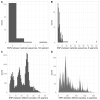
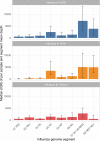
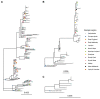
Results: From 941 infections, successful sequencing was achieved in 292/388 (75 %) available Oxfordshire samples: 231 (79 %) A/H3N2, 53 (18 %) A/H1N1, and 8 (3 %) B/Victoria and in 53/113 (47 %) UK-wide samples. Sequencing was more successful at lower Ct values. Most same-sample replicate sequences had identical haemagglutinin segments (124/141, 88 %); 36/39 (92 %) Illumina vs. Nanopore comparisons were identical, and 3 (8 %) differed by 1 variant. Comparison of Oxfordshire and UK-wide sequences showed frequent inter-regional transmission. Infections were closely-related to 2022-23 vaccine strains. Only one sample had a neuraminidase inhibitor resistance mutation. 849/941 (90 %) Oxfordshire infections were community-acquired. 63/88 (72 %) potentially healthcare-associated cases shared a hospital ward with ≥ 1 known infectious case. 33 epidemiologically-plausible transmission links had sequencing data for both source and recipient: 8 were within ≤ 5 SNPs, of these, 5 (63 %) involved potential sources that were also hospital-acquired.
Conclusions: Nanopore influenza sequencing was reproducible and antiviral resistance rare. Inter-regional transmission was common; most infections were genomically similar. Hospital-acquired infections are likely an important source of nosocomial transmission and should be prioritised for infection prevention and control.
Keywords: Epidemiology; Influenza; Respiratory virus; Sequencing.
Copyright © 2024 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest No author has a conflict of interest to declare.
Figures

References
-
- UK Health Security Agency. Surveillance of influenza and other seasonal respiratory viruses in the UK, winter 2022 to 2023; 2023. 〈https://www.gov.uk/government/statistics/annual-flu-reports/surveillance...〉 [Accessed Oct 31, 2023].
-
- UK Health Security Agency. Guidance on use of antiviral agents for the treatment and prophylaxis of seasonal influenza; 2021. 〈https://www.gov.uk/government/publications/influenza-treatment-and-proph...〉 [Accessed Oct 31, 2023].
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

