The impact of exercise on gene regulation in association with complex trait genetics
- PMID: 38693125
- PMCID: PMC11063075
- DOI: 10.1038/s41467-024-45966-w
The impact of exercise on gene regulation in association with complex trait genetics
Abstract
Endurance exercise training is known to reduce risk for a range of complex diseases. However, the molecular basis of this effect has been challenging to study and largely restricted to analyses of either few or easily biopsied tissues. Extensive transcriptome data collected across 15 tissues during exercise training in rats as part of the Molecular Transducers of Physical Activity Consortium has provided a unique opportunity to clarify how exercise can affect tissue-specific gene expression and further suggest how exercise adaptation may impact complex disease-associated genes. To build this map, we integrate this multi-tissue atlas of gene expression changes with gene-disease targets, genetic regulation of expression, and trait relationship data in humans. Consensus from multiple approaches prioritizes specific tissues and genes where endurance exercise impacts disease-relevant gene expression. Specifically, we identify a total of 5523 trait-tissue-gene triplets to serve as a valuable starting point for future investigations [Exercise; Transcription; Human Phenotypic Variation].
© 2024. The Author(s).
Conflict of interest statement
S.B.M. is a consultant for BioMarin, MyOme and Tenaya Therapeutics. These companies are broadly interested in treatments for rare and common genetic diseases but had no input on any component of this study. The authors have no other competing interests to declare.
Figures

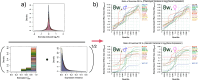


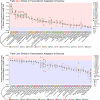

References
-
- Fiuza-Luces C, et al. Exercise benefits in cardiovascular disease: beyond attenuation of traditional risk factors. Nat. Rev. Cardiol. 2018;15:731–743. - PubMed
-
- MoTrPAC Study Group Temporal dynamics of the multi-omic response to endurance exercise training across tissues. Preprint at https://www.biorxiv.org/content/10.1101/2022.09.21.508770v2 (2022). - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- U01 AG055135/AG/NIA NIH HHS/United States
- U01 AR071158/AR/NIAMS NIH HHS/United States
- U01 AR071130/AR/NIAMS NIH HHS/United States
- U01 AR071133/AR/NIAMS NIH HHS/United States
- U24 DK112340/DK/NIDDK NIH HHS/United States
- P30 DK036836/DK/NIDDK NIH HHS/United States
- P30 AG044271/AG/NIA NIH HHS/United States
- P20 GM144269/GM/NIGMS NIH HHS/United States
- U24 OD026629/OD/NIH HHS/United States
- U24 DK112342/DK/NIDDK NIH HHS/United States
- U24 DK112331/DK/NIDDK NIH HHS/United States
- U01 AG070960/AG/NIA NIH HHS/United States
- U24 AR071113/AR/NIAMS NIH HHS/United States
- U01 AR071160/AR/NIAMS NIH HHS/United States
- U24 DK112348/DK/NIDDK NIH HHS/United States
- U01 AG055137/AG/NIA NIH HHS/United States
- U01 AG070959/AG/NIA NIH HHS/United States
- U01 AG055133/AG/NIA NIH HHS/United States
- U01 AR071128/AR/NIAMS NIH HHS/United States
- U01 AR071124/AR/NIAMS NIH HHS/United States
- U01 AG070928/AG/NIA NIH HHS/United States
- T15 LM007033/LM/NLM NIH HHS/United States
- U24 DK112341/DK/NIDDK NIH HHS/United States
- U24 DK112349/DK/NIDDK NIH HHS/United States
- U01 AR071150/AR/NIAMS NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- U24 DK112326/DK/NIDDK NIH HHS/United States

