A novel GH3-β-glucosidase from soda lake metagenomic libraries with desirable properties for biomass degradation
- PMID: 38693138
- PMCID: PMC11063200
- DOI: 10.1038/s41598-024-60645-y
A novel GH3-β-glucosidase from soda lake metagenomic libraries with desirable properties for biomass degradation
Abstract
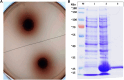
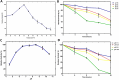
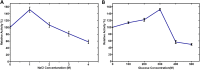
Beta-glucosidases catalyze the hydrolysis of the glycosidic bonds of cellobiose, producing glucose, which is a rate-limiting step in cellulose biomass degradation. In industrial processes, β-glucosidases that are tolerant to glucose and stable under harsh industrial reaction conditions are required for efficient cellulose hydrolysis. In this study, we report the molecular cloning, Escherichia coli expression, and functional characterization of a β-glucosidase from the gene, CelGH3_f17, identified from metagenomics libraries of an Ethiopian soda lake. The CelGH3_f17 gene sequence contains a glycoside hydrolase family 3 catalytic domain (GH3). The heterologous expressed and purified enzyme exhibited optimal activity at 50 °C and pH 8.5. In addition, supplementation of 1 M salt and 300 mM glucose enhanced the β-glucosidase activity. Most of the metal ions and organic solvents tested did not affect the β-glucosidase activity. However, Cu2+ and Mn2+ ions, Mercaptoethanol and Triton X-100 reduce the activity of the enzyme. The studied β-glucosidase enzyme has multiple industrially desirable properties including thermostability, and alkaline, salt, and glucose tolerance.
Keywords: Beta-glucosidase; Enzyme characterisation; Glycoside hydrolase family 3 (GH3); Soda lakes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Overexpression and characterization of a glucose-tolerant β-glucosidase from T. aotearoense with high specific activity for cellobiose.Appl Microbiol Biotechnol. 2015 Nov;99(21):8903-15. doi: 10.1007/s00253-015-6619-9. Epub 2015 May 9. Appl Microbiol Biotechnol. 2015. PMID: 25957152
-
A novel β-glucosidase from a hot-spring metagenome shows elevated thermal stability and tolerance to glucose and ethanol.Enzyme Microb Technol. 2021 Apr;145:109764. doi: 10.1016/j.enzmictec.2021.109764. Epub 2021 Feb 11. Enzyme Microb Technol. 2021. PMID: 33750538
-
Biochemical and Structural Characterization of a Highly Glucose-Tolerant β-Glucosidase from the Termite Reticulitermes perilucifugus.Int J Mol Sci. 2025 Mar 28;26(7):3118. doi: 10.3390/ijms26073118. Int J Mol Sci. 2025. PMID: 40243882 Free PMC article.
-
β-Glucosidase from the hyperthermophilic archaeon Thermococcus sp. is a salt-tolerant enzyme that is stabilized by its reaction product glucose.Appl Microbiol Biotechnol. 2016 Oct;100(19):8399-409. doi: 10.1007/s00253-016-7601-x. Epub 2016 May 19. Appl Microbiol Biotechnol. 2016. PMID: 27198723
-
Recent Advances in β-Glucosidase Sequence and Structure Engineering: A Brief Review.Molecules. 2023 Jun 25;28(13):4990. doi: 10.3390/molecules28134990. Molecules. 2023. PMID: 37446652 Free PMC article. Review.
Cited by
-
Characterization, thermostable mechanism, and molecular docking of a novel glucose-tolerant β-glucosidase/β-galactosidase from the GH1 family isolated from Rehai hot spring.Front Microbiol. 2025 Apr 11;16:1559242. doi: 10.3389/fmicb.2025.1559242. eCollection 2025. Front Microbiol. 2025. PMID: 40291800 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

