Airway hillocks are injury-resistant reservoirs of unique plastic stem cells
- PMID: 38693267
- PMCID: PMC11890216
- DOI: 10.1038/s41586-024-07377-1
Airway hillocks are injury-resistant reservoirs of unique plastic stem cells
Abstract
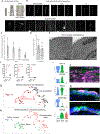
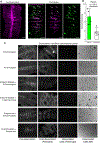
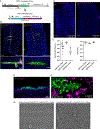
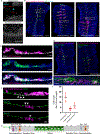
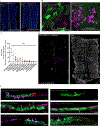
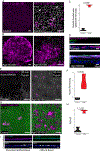
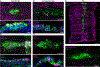
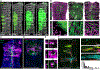
Airway hillocks are stratified epithelial structures of unknown function1. Hillocks persist for months and have a unique population of basal stem cells that express genes associated with barrier function and cell adhesion. Hillock basal stem cells continually replenish overlying squamous barrier cells. They exhibit dramatically higher turnover than the abundant, largely quiescent classic pseudostratified airway epithelium. Hillocks resist a remarkably broad spectrum of injuries, including toxins, infection, acid and physical injury because hillock squamous cells shield underlying hillock basal stem cells from injury. Hillock basal stem cells are capable of massive clonal expansion that is sufficient to resurface denuded airway, and eventually regenerate normal airway epithelium with each of its six component cell types. Hillock basal stem cells preferentially stratify and keratinize in the setting of retinoic acid signalling inhibition, a known cause of squamous metaplasia2,3. Here we show that mouse hillock expansion is the cause of vitamin A deficiency-induced squamous metaplasia. Finally, we identify human hillocks whose basal stem cells generate functional squamous barrier structures in culture. The existence of hillocks reframes our understanding of airway epithelial regeneration. Furthermore, we show that hillocks are one origin of 'squamous metaplasia', which is long thought to be a precursor of lung cancer.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures






References
-
- Lancillotti F, Darwiche N, Celli G & De Luca LM Retinoid status and the control of keratin expression and adhesion during the histogenesis of squamous metaplasia of tracheal epithelium. Cancer Res. 52, 6144–6152 (1992). - PubMed
-
- Chopra DP Retinoid reversal of squamous metaplasia in organ cultures of tracheas derived from hamsters fed on vitamin A-deficient diet. Eur. J. Cancer Clin. Oncol. 19, 847–857 (1983). - PubMed
MeSH terms
Substances
Grants and funding
- R01 HL118185/HL/NHLBI NIH HHS/United States
- U24 HL148865/HL/NHLBI NIH HHS/United States
- UH3 CA268117/CA/NCI NIH HHS/United States
- F31 HL165736/HL/NHLBI NIH HHS/United States
- UG3 CA268117/CA/NCI NIH HHS/United States
- K08 HL124298/HL/NHLBI NIH HHS/United States
- P30 DK057521/DK/NIDDK NIH HHS/United States
- T32 HL116275/HL/NHLBI NIH HHS/United States
- R01 HL157221/HL/NHLBI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- P30 DK135043/DK/NIDDK NIH HHS/United States
- R01 HL142559/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

