Multimodal decoding of human liver regeneration
- PMID: 38693268
- PMCID: PMC11153152
- DOI: 10.1038/s41586-024-07376-2
Multimodal decoding of human liver regeneration
Abstract
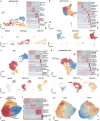
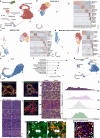
The liver has a unique ability to regenerate1,2; however, in the setting of acute liver failure (ALF), this regenerative capacity is often overwhelmed, leaving emergency liver transplantation as the only curative option3-5. Here, to advance understanding of human liver regeneration, we use paired single-nucleus RNA sequencing combined with spatial profiling of healthy and ALF explant human livers to generate a single-cell, pan-lineage atlas of human liver regeneration. We uncover a novel ANXA2+ migratory hepatocyte subpopulation, which emerges during human liver regeneration, and a corollary subpopulation in a mouse model of acetaminophen (APAP)-induced liver regeneration. Interrogation of necrotic wound closure and hepatocyte proliferation across multiple timepoints following APAP-induced liver injury in mice demonstrates that wound closure precedes hepatocyte proliferation. Four-dimensional intravital imaging of APAP-induced mouse liver injury identifies motile hepatocytes at the edge of the necrotic area, enabling collective migration of the hepatocyte sheet to effect wound closure. Depletion of hepatocyte ANXA2 reduces hepatocyte growth factor-induced human and mouse hepatocyte migration in vitro, and abrogates necrotic wound closure following APAP-induced mouse liver injury. Together, our work dissects unanticipated aspects of liver regeneration, demonstrating an uncoupling of wound closure and hepatocyte proliferation and uncovering a novel migratory hepatocyte subpopulation that mediates wound closure following liver injury. Therapies designed to promote rapid reconstitution of normal hepatic microarchitecture and reparation of the gut-liver barrier may advance new areas of therapeutic discovery in regenerative medicine.
© 2024. The Author(s).
Conflict of interest statement
N.C.H. has received research funding from AbbVie, Pfizer, Gilead, Boehringer-Ingelheim and Galecto, and is an advisor or consultant for AstraZeneca, GSK, MSD, Galecto and Pliant Therapeutics. R.J. is an inventor of OPA (licensed to Mallinckrodt Pharma) and founder of Yaqrit Discovery, Hepyx Limited and Cyberliver. All other authors declare no competing interests.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

