Targeted desialylation and cytolysis of tumour cells by fusing a sialidase to a bispecific T-cell engager
- PMID: 38693431
- PMCID: PMC11577304
- DOI: 10.1038/s41551-024-01202-w
Targeted desialylation and cytolysis of tumour cells by fusing a sialidase to a bispecific T-cell engager
Abstract
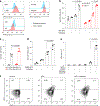
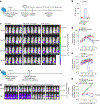
Bispecific T-cell engagers (BiTEs) bring together tumour cells and cytotoxic T cells by binding to specific cell-surface tumour antigens and T-cell receptors, and have been clinically successful for the treatment of B-cell malignancies. Here we show that a BiTE-sialidase fusion protein enhances the susceptibility of solid tumours to BiTE-mediated cytolysis of tumour cells via targeted desialylation-that is, the removal of terminal sialic acid residues on glycans-at the BiTE-induced T-cell-tumour-cell interface. In xenograft and syngeneic mouse models of leukaemia and of melanoma and breast cancer, and compared with the parental BiTE molecules, targeted desialylation via the BiTE-sialidase fusion proteins enhanced the formation of immunological synapses, T-cell activation and T-cell-mediated tumour-cell cytolysis in the presence of the target antigen. The targeted desialylation of tumour cells may enhance the potency of therapies relying on T-cell engagers.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures





References
-
- Goebeler M-E & Bargou RC T cell-engaging therapies—BiTEs and beyond. Nat. Rev. Clin. Oncol---. 17, 418–434 (2020). - PubMed
-
- Przepiorka D. et al. FDA approval: blinatumomab. Clin. Cancer Res. 21, 4035–4039 (2015). - PubMed
-
- Arvedson T. et al. Targeting solid tumors with bispecific T cell engager immune therapy. Annu. Rev. Cancer Biol. 6, 17–34 (2021).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

