18S rDNA sequence-structure phylogeny of the eukaryotes simultaneously inferred from sequences and their individual secondary structures
- PMID: 38693573
- PMCID: PMC11064340
- DOI: 10.1186/s13104-024-06786-9
18S rDNA sequence-structure phylogeny of the eukaryotes simultaneously inferred from sequences and their individual secondary structures
Abstract
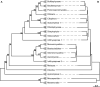
Objective: The eukaryotic tree of life has been subject of numerous studies ever since the nineteenth century, with more supergroups and their sister relations being decoded in the last years. In this study, we reconstructed the phylogeny of eukaryotes using complete 18S rDNA sequences and their individual secondary structures simultaneously. After the sequence-structure data was encoded, it was automatically aligned and analyzed using sequence-only as well as sequence-structure approaches. We present overall neighbor-joining trees of 211 eukaryotes as well as the respective profile neighbor-joining trees, which helped to resolve the basal branching pattern. A manually chosen subset was further inspected using neighbor-joining, maximum parsimony, and maximum likelihood analyses. Additionally, the 75 and 100 percent consensus structures of the subset were predicted.
Results: All sequence-structure approaches show improvements compared to the respective sequence-only approaches: the average bootstrap support per node of the sequence-structure profile neighbor-joining analyses with 90.3, was higher than the average bootstrap support of the sequence-only profile neighbor-joining analysis with 73.9. Also, the subset analyses using sequence-structure data were better supported. Furthermore, more subgroups of the supergroups were recovered as monophyletic and sister group relations were much more comparable to results as obtained by multi-marker analyses.
Keywords: 18S rDNA; Eukaryotes; Phylogenetics; Secondary structure.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

