MiR-29 and MiR-140 regulate TRAIL-induced drug tolerance in lung cancer
- PMID: 38693921
- PMCID: PMC11062278
- DOI: 10.1080/19768354.2024.2345644
MiR-29 and MiR-140 regulate TRAIL-induced drug tolerance in lung cancer
Abstract
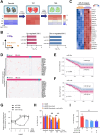
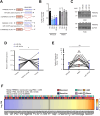
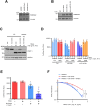
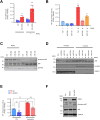
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) has chemotherapeutic potential as a regulator of an extrinsic apoptotic ligand, but its effect as a drug is limited by innate and acquired resistance. Recent findings suggest that an intermediate drug tolerance could mediate acquired resistance, which has made the main obstacle for limited utility of TRAIL as an anti-cancer therapeutics. We propose miRNA-dependent epigenetic modification drives the drug tolerant state in TRAIL-induced drug tolerant (TDT). Transcriptomic analysis revealed miR-29 target gene activation in TDT cells, showing oncogenic signature in lung cancer. Also, the restored TRAIL-sensitivity was associated with miR-29ac and 140-5p expressions, which is known as tumor suppressor by suppressing oncogenic protein RSK2 (p90 ribosomal S6 kinase), further confirmed in patient samples. Moreover, we extended this finding into 119 lung cancer cell lines from public data set, suggesting a significant correlation between TRAIL-sensitivity and RSK2 mRNA expression. Finally, we found that increased RSK2 mRNA is responsible for NF-κB activation, which we previously showed as a key determinant in both innate and acquired TRAIL-resistance. Our findings support further investigation of miR-29ac and -140-5p inhibition to maintain TRAIL-sensitivity and improve the durability of response to TRAIL in lung cancer.
Keywords: MicroRNAs; RSK2; TRAIL-persistence; lung cancer.
© 2024 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
Metformin Causes G1-Phase Arrest via Down-Regulation of MiR-221 and Enhances TRAIL Sensitivity through DR5 Up-Regulation in Pancreatic Cancer Cells.PLoS One. 2015 May 8;10(5):e0125779. doi: 10.1371/journal.pone.0125779. eCollection 2015. PLoS One. 2015. PMID: 25955843 Free PMC article.
-
miR-221 Augments TRAIL-Mediated Apoptosis in Prostate Cancer Cells by Inducing Endogenous TRAIL Expression and Targeting the Functional Repressors SOCS3 and PIK3R1.Biomed Res Int. 2019 Nov 14;2019:6392748. doi: 10.1155/2019/6392748. eCollection 2019. Biomed Res Int. 2019. PMID: 31828111 Free PMC article.
-
Tumor necrosis factor-related apoptosis-inducing ligand induces the expression of proinflammatory cytokines in macrophages and re-educates tumor-associated macrophages to an antitumor phenotype.Mol Biol Cell. 2015 Sep 15;26(18):3178-89. doi: 10.1091/mbc.E15-04-0209. Epub 2015 Jul 29. Mol Biol Cell. 2015. PMID: 26224317 Free PMC article.
-
Regulation of sensitivity to TRAIL by the PTEN tumor suppressor.Vitam Horm. 2004;67:409-26. doi: 10.1016/S0083-6729(04)67021-X. Vitam Horm. 2004. PMID: 15110188 Review.
-
Genetic and Epigenetic Regulation of the Innate Immune Response to Gout.Immunol Invest. 2023 Apr;52(3):364-397. doi: 10.1080/08820139.2023.2168554. Epub 2023 Feb 6. Immunol Invest. 2023. PMID: 36745138 Review.
References
-
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. . 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't]. Nature. 483(7391):603–607. eng. doi:10.1038/nature11003. - DOI - PMC - PubMed
-
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME.. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms [Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, P.H.S.]. Science. 286(5443):1358–1362. eng. doi:10.1126/science.286.5443.1358. - DOI - PubMed
-
- Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L.. 1997. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway [Research Support, Non-U.S. Gov't]. Immunity. 7(6):821–830. eng. doi:10.1016/S1074-7613(00)80400-8. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
