Variations in the persistence of 5'-end genomic and subgenomic SARS-CoV-2 RNAs in wastewater from aircraft, airports and wastewater treatment plants
- PMID: 38694057
- PMCID: PMC11061675
- DOI: 10.1016/j.heliyon.2024.e29703
Variations in the persistence of 5'-end genomic and subgenomic SARS-CoV-2 RNAs in wastewater from aircraft, airports and wastewater treatment plants
Abstract
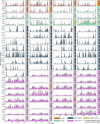
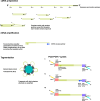
Wastewater sequencing has become a powerful supplement to clinical testing in monitoring SARS-CoV-2 infections in the post-COVID-19 pandemic era. While its applications in measuring the viral burden and main circulating lineages in the community have proved their efficacy, the variations in sequencing quality and coverage across the different regions of the SARS-CoV-2 genome are not well understood. Furthermore, it is unclear how different sample origins, viral extraction and concentration methods and environmental factors impact the reads sequenced from wastewater. Using high-coverage, amplicon-based, paired-end read sequencing of viral RNA extracted from wastewater collected directly from aircraft, pooled from different aircraft and airport buildings or from regular wastewater plants, we assessed the genome coverage across the sample groups with a focus on the 5'-end region covering the leader sequence and investigated whether it was possible to detect subgenomic RNA from viral material recovered from wastewater. We identified distinct patterns in the persistence of the different genomic regions across the different types of wastewaters and the existence of chimeric reads mapping to non-amplified regions. Our findings suggest that preservation of the 5'-end of the genome and the ability to detect subgenomic RNA reads, though highly susceptible to environment and sample processing conditions, may be indicative of the quality and amount of the viral RNA present in wastewater.
Keywords: SARS-CoV-2; Subgenomic RNA; Transcription/replication; Wastewater.
© 2024 Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Unambiguous detection of SARS-CoV-2 subgenomic mRNAs with single-cell RNA sequencing.Microbiol Spectr. 2023 Sep 7;11(5):e0077623. doi: 10.1128/spectrum.00776-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37676044 Free PMC article.
-
Genomic surveillance of Canadian airport wastewater samples allows early detection of emerging SARS-CoV-2 lineages.Sci Rep. 2024 Nov 3;14(1):26534. doi: 10.1038/s41598-024-76925-6. Sci Rep. 2024. PMID: 39489759 Free PMC article.
-
Intensity of sample processing methods impacts wastewater SARS-CoV-2 whole genome amplicon sequencing outcomes.Sci Total Environ. 2023 Jun 10;876:162572. doi: 10.1016/j.scitotenv.2023.162572. Epub 2023 Mar 4. Sci Total Environ. 2023. PMID: 36871720 Free PMC article.
-
A review on detection of SARS-CoV-2 RNA in wastewater in light of the current knowledge of treatment process for removal of viral fragments.J Environ Manage. 2021 Dec 1;299:113563. doi: 10.1016/j.jenvman.2021.113563. Epub 2021 Aug 19. J Environ Manage. 2021. PMID: 34488114 Free PMC article. Review.
-
A State-of-the-Art Scoping Review on SARS-CoV-2 in Sewage Focusing on the Potential of Wastewater Surveillance for the Monitoring of the COVID-19 Pandemic.Food Environ Virol. 2022 Dec;14(4):315-354. doi: 10.1007/s12560-021-09498-6. Epub 2021 Nov 2. Food Environ Virol. 2022. PMID: 34727334 Free PMC article.
Cited by
-
Magnetic Carbon Bead-Based Concentration Method for SARS-CoV-2 Detection in Wastewater.Food Environ Virol. 2024 Dec 31;17(1):8. doi: 10.1007/s12560-024-09623-1. Food Environ Virol. 2024. PMID: 39741220
References
-
- Toledo D.M., Robbins A.A., Gallagher T.L., Hershberger K.C., Barney R.E., Salmela S.M., Pilcher D., Cervinski M.A., Nerenz R.D., Szczepiorkowski Z.M., Tsongalis G.J., Lefferts J.A., Martin I.W., Hubbard J.A. Wastewater-based SARS-CoV-2 surveillance in northern new England. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.02207-21. - DOI - PMC - PubMed
-
- Vallejo J.A., Trigo-Tasende N., Rumbo-Feal S., Conde-Pérez K., López-Oriona Á., Barbeito I., Vaamonde M., Tarrío-Saavedra J., Reif R., Ladra S., Rodiño-Janeiro B.K., Nasser-Ali M., Cid Á., Veiga M., Acevedo A., Lamora C., Bou G., Cao R., Poza M. Modeling the number of people infected with SARS-COV-2 from wastewater viral load in Northwest Spain. Sci. Total Environ. 2022;811 doi: 10.1016/j.scitotenv.2021.152334. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

