Activation of immune pathways in common bed bugs, Cimex lectularius, in response to bacterial immune challenges - a transcriptomics analysis
- PMID: 38694504
- PMCID: PMC11061471
- DOI: 10.3389/fimmu.2024.1384193
Activation of immune pathways in common bed bugs, Cimex lectularius, in response to bacterial immune challenges - a transcriptomics analysis
Abstract
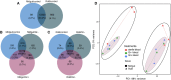
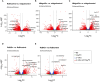
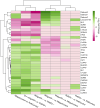
The common bed bug, Cimex lectularius, is an urban pest of global health significance, severely affecting the physical and mental health of humans. In contrast to most other blood-feeding arthropods, bed bugs are not major vectors of pathogens, but the underlying mechanisms for this phenomenon are largely unexplored. Here, we present the first transcriptomics study of bed bugs in response to immune challenges. To study transcriptional variations in bed bugs following ingestion of bacteria, we extracted and processed mRNA from body tissues of adult male bed bugs after ingestion of sterile blood or blood containing the Gram-positive (Gr+) bacterium Bacillus subtilis or the Gram-negative (Gr-) bacterium Escherichia coli. We analyzed mRNA from the bed bugs' midgut (the primary tissue involved in blood ingestion) and from the rest of their bodies (RoB; body minus head and midgut tissues). We show that the midgut exhibits a stronger immune response to ingestion of bacteria than the RoB, as indicated by the expression of genes encoding antimicrobial peptides (AMPs). Both the Toll and Imd signaling pathways, associated with immune responses, were highly activated by the ingestion of bacteria. Bacterial infection in bed bugs further provides evidence for metabolic reconfiguration and resource allocation in the bed bugs' midgut and RoB to promote production of AMPs. Our data suggest that infection with particular pathogens in bed bugs may be associated with altered metabolic pathways within the midgut and RoB that favors immune responses. We further show that multiple established cellular immune responses are preserved and are activated by the presence of specific pathogens. Our study provides a greater understanding of nuances in the immune responses of bed bugs towards pathogens that ultimately might contribute to novel bed bug control tactics.
Keywords: antimicrobial peptides; bed bugs; immune transcriptomics; immunization; innate immunity; toll & Imd pathways; vectors & pathogens.
Copyright © 2024 Meraj, Salcedo-Porras, Lowenberger and Gries.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A novel prolixicin identified in common bed bugs with activity against both bacteria and parasites.Sci Rep. 2024 Jun 15;14(1):13818. doi: 10.1038/s41598-024-64691-4. Sci Rep. 2024. PMID: 38879638 Free PMC article.
-
Time- and tissue-specific antimicrobial activity of the common bed bug in response to blood feeding and immune activation by bacterial injection.J Insect Physiol. 2021 Nov-Dec;135:104322. doi: 10.1016/j.jinsphys.2021.104322. Epub 2021 Oct 10. J Insect Physiol. 2021. PMID: 34644597
-
Characterization of New Defensin Antimicrobial Peptides and Their Expression in Bed Bugs in Response to Bacterial Ingestion and Injection.Int J Mol Sci. 2022 Sep 29;23(19):11505. doi: 10.3390/ijms231911505. Int J Mol Sci. 2022. PMID: 36232802 Free PMC article.
-
Bed bugs (Cimex lectularius) and clinical consequences of their bites.JAMA. 2009 Apr 1;301(13):1358-66. doi: 10.1001/jama.2009.405. JAMA. 2009. PMID: 19336711 Review.
-
Historical and Contemporary Control Options Against Bed Bugs, Cimex spp.Annu Rev Entomol. 2023 Jan 23;68:169-190. doi: 10.1146/annurev-ento-120220-015010. Epub 2022 Oct 5. Annu Rev Entomol. 2023. PMID: 36198396 Review.
Cited by
-
Were bed bugs the first urban pest insect? Genome-wide patterns of bed bug demography mirror global human expansion.Biol Lett. 2025 May;21(5):20250061. doi: 10.1098/rsbl.2025.0061. Epub 2025 May 28. Biol Lett. 2025. PMID: 40425045
-
Functionality of highly diverged Imd like genes identified in stinkbugs and bedbugs.Sci Rep. 2025 Jun 4;15(1):19520. doi: 10.1038/s41598-025-02641-4. Sci Rep. 2025. PMID: 40467703 Free PMC article.
-
Characterization of temporal expression of immune genes in female locust challenged by fungal pathogen, Aspergillus sp.Front Immunol. 2025 Apr 28;16:1565964. doi: 10.3389/fimmu.2025.1565964. eCollection 2025. Front Immunol. 2025. PMID: 40356898 Free PMC article.
-
A novel prolixicin identified in common bed bugs with activity against both bacteria and parasites.Sci Rep. 2024 Jun 15;14(1):13818. doi: 10.1038/s41598-024-64691-4. Sci Rep. 2024. PMID: 38879638 Free PMC article.
-
Common Bed Bugs: Non-Viable Hosts for Trypanosoma rangeli Parasites.Cells. 2024 Dec 11;13(24):2042. doi: 10.3390/cells13242042. Cells. 2024. PMID: 39768134 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

