Ocular surface immune transcriptome and tear cytokines in corneal infection patients
- PMID: 38694515
- PMCID: PMC11061372
- DOI: 10.3389/fcimb.2024.1346821
Ocular surface immune transcriptome and tear cytokines in corneal infection patients
Abstract
Background: Microbial keratitis is one of the leading causes of blindness globally. An overactive immune response during an infection can exacerbate damage, causing corneal opacities and vision loss. This study aimed to identify the differentially expressed genes between corneal infection patients and healthy volunteers within the cornea and conjunctiva and elucidate the contributing pathways to these conditions' pathogenesis. Moreover, it compared the corneal and conjunctival transcriptomes in corneal-infected patients to cytokine levels in tears.
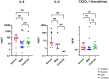
Methods: Corneal and conjunctival swabs were collected from seven corneal infection patients and three healthy controls under topical anesthesia. RNA from seven corneal infection patients and three healthy volunteers were analyzed by RNA sequencing (RNA-Seq). Tear proteins were extracted from Schirmer strips via acetone precipitation from 38 cases of corneal infection and 14 healthy controls. The cytokines and chemokines IL-1β, IL-6, CXCL8 (IL-8), CX3CL1, IL-10, IL-12 (p70), IL-17A, and IL-23 were measured using an antibody bead assay.
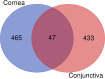
Results: A total of 512 genes were found to be differentially expressed in infected corneas compared to healthy corneas, with 508 being upregulated and four downregulated (fold-change (FC) <-2 or > 2 and adjusted p <0.01). For the conjunctiva, 477 were upregulated, and 3 were downregulated (FC <-3 or ≥ 3 and adjusted p <0.01). There was a significant overlap in cornea and conjunctiva gene expression in patients with corneal infections. The genes were predominantly associated with immune response, regulation of angiogenesis, and apoptotic signaling pathways. The most highly upregulated gene was CXCL8 (which codes for IL-8 protein). In patients with corneal infections, the concentration of IL-8 protein in tears was relatively higher in patients compared to healthy controls but did not show statistical significance.
Conclusions: During corneal infection, many genes were upregulated, with most of them being associated with immune response, regulation of angiogenesis, and apoptotic signaling. The findings may facilitate the development of treatments for corneal infections that can dampen specific aspects of the immune response to reduce scarring and preserve sight.
Keywords: bacteria; conjunctiva; corneal infection; gene expression; keratitis; ocular surface; transcriptome.
Copyright © 2024 Alenezi, Parnell, Schibeci, Ozkan, Willcox, White and Carnt.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Persistence of Innate Immune Pathways in Late Stage Human Bacterial and Fungal Keratitis: Results from a Comparative Transcriptome Analysis.Front Cell Infect Microbiol. 2017 May 18;7:193. doi: 10.3389/fcimb.2017.00193. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28573109 Free PMC article.
-
Altered cytokine balance in the tear fluid and conjunctiva of patients with Sjögren's syndrome keratoconjunctivitis sicca.Curr Eye Res. 1999 Sep;19(3):201-11. doi: 10.1076/ceyr.19.3.201.5309. Curr Eye Res. 1999. PMID: 10487957
-
Correlation between human tear cytokine levels and cellular corneal changes in patients with bacterial keratitis by in vivo confocal microscopy.Invest Ophthalmol Vis Sci. 2014 Oct 16;55(11):7457-66. doi: 10.1167/iovs.14-15411. Invest Ophthalmol Vis Sci. 2014. PMID: 25324281 Free PMC article.
-
Bilateral Alterations in Corneal Nerves, Dendritic Cells, and Tear Cytokine Levels in Ocular Surface Disease.Cornea. 2016 Nov;35 Suppl 1(Suppl 1):S65-S70. doi: 10.1097/ICO.0000000000000989. Cornea. 2016. PMID: 27617877 Free PMC article. Review.
-
Differential expression of tear film cytokines in Stevens-Johnson syndrome patients and comparative review of literature.Sci Rep. 2021 Sep 16;11(1):18433. doi: 10.1038/s41598-021-97575-y. Sci Rep. 2021. PMID: 34531438 Free PMC article. Review.
Cited by
-
Nigericin Induces Paraptosis-Like Cell Death Instead of Pyroptosis in Corneal Keratocytes.FASEB J. 2025 Jun 30;39(12):e70740. doi: 10.1096/fj.202500502R. FASEB J. 2025. PMID: 40540302 Free PMC article.
-
Fusarium spp. and Aspergillus flavus infection induces pathogen-specific and pathogen-independent host immune response in patients with fungal keratitis.Front Cell Infect Microbiol. 2025 May 30;15:1560628. doi: 10.3389/fcimb.2025.1560628. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40521031 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

