The complete genome sequence of unculturable Mycoplasma faucium obtained through clinical metagenomic next-generation sequencing
- PMID: 38694516
- PMCID: PMC11062135
- DOI: 10.3389/fcimb.2024.1368923
The complete genome sequence of unculturable Mycoplasma faucium obtained through clinical metagenomic next-generation sequencing
Abstract
Introduction: Diagnosing Mycoplasma faucium poses challenges, and it's unclear if its rare isolation is due to infrequent occurrence or its fastidious nutritional requirements.
Methods: This study analyzes the complete genome sequence of M. faucium, obtained directly from the pus of a sternum infection in a lung transplant patient using metagenomic sequencing.
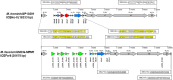
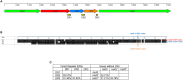
Results: Genome analysis revealed limited therapeutic options for the M. faucium infection, primarily susceptibility to tetracyclines. Three classes of mobile genetic elements were identified: two new insertion sequences, a new prophage (phiUMCG-1), and a species-specific variant of a mycoplasma integrative and conjugative element (MICE). Additionally, a Type I Restriction-Modification system was identified, featuring 5'-terminally truncated hsdS pseudogenes with overlapping repeats, indicating the potential for forming alternative hsdS variants through recombination.
Conclusion: This study represents the first-ever acquisition of a complete circularized bacterial genome directly from a patient sample obtained from invasive infection of a primary sterile site using culture-independent, PCR-free clinical metagenomics.
Keywords: Mycoplasma faucium; ice; insertion sequence; lung transplant patient; metagenomics.
Copyright © 2024 Sabat, Durfee, Baldwin, Akkerboom, Voss, Friedrich and Bathoorn.
Conflict of interest statement
Authors TD and SB are employees of the company DNASTAR, Inc. SB is a minor shareholder in DNASTAR, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

