The impact of antibiotic exposure on antibiotic resistance gene dynamics in the gut microbiota of inflammatory bowel disease patients
- PMID: 38694799
- PMCID: PMC11061493
- DOI: 10.3389/fmicb.2024.1382332
The impact of antibiotic exposure on antibiotic resistance gene dynamics in the gut microbiota of inflammatory bowel disease patients
Abstract
Background: While antibiotics are commonly used to treat inflammatory bowel disease (IBD), their widespread application can disturb the gut microbiota and foster the emergence and spread of antibiotic resistance. However, the dynamic changes to the human gut microbiota and direction of resistance gene transmission under antibiotic effects have not been clearly elucidated.
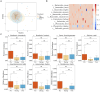
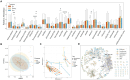
Methods: Based on the Human Microbiome Project, a total of 90 fecal samples were collected from 30 IBD patients before, during and after antibiotic treatment. Through the analysis workflow of metagenomics, we described the dynamic process of changes in bacterial communities and resistance genes pre-treatment, during and post-treatment. We explored potential consistent relationships between gut microbiota and resistance genes, and established gene transmission networks among species before and after antibiotic use.
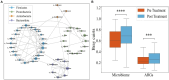
Results: Exposure to antibiotics can induce alterations in the composition of the gut microbiota in IBD patients, particularly a reduction in probiotics, which gradually recovers to a new steady state after cessation of antibiotics. Network analyses revealed intra-phylum transfers of resistance genes, predominantly between taxonomically close organisms. Specific resistance genes showed increased prevalence and inter-species mobility after antibiotic cessation.
Conclusion: This study demonstrates that antibiotics shape the gut resistome through selective enrichment and promotion of horizontal gene transfer. The findings provide insights into ecological processes governing resistance gene dynamics and dissemination upon antibiotic perturbation of the microbiota. Optimizing antibiotic usage may help limit unintended consequences like increased resistance in gut bacteria during IBD management.
Keywords: antibiotics resistance genes; drug resistance; gut microbiome; horizontal gene transfer; metagenomics.
Copyright © 2024 Zhang, Xue, Wang, Zhang, Xu and Yu.
Conflict of interest statement
GX and LX were employed by the company Hotgen Biotech Co., Ltd. FW and LX was also employed by Beijing YuGen Pharmaceutical Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Antibiotic resistance gene sharing networks and the effect of dietary nutritional content on the canine and feline gut resistome.Anim Microbiome. 2020 Feb 7;2(1):4. doi: 10.1186/s42523-020-0022-2. Anim Microbiome. 2020. PMID: 33500005 Free PMC article.
-
The human gut resistome.Philos Trans R Soc Lond B Biol Sci. 2015 Jun 5;370(1670):20140087. doi: 10.1098/rstb.2014.0087. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 25918444 Free PMC article. Review.
-
The effect of antibiotics on the gut microbiome: a metagenomics analysis of microbial shift and gut antibiotic resistance in antibiotic treated mice.BMC Genomics. 2020 Mar 30;21(1):263. doi: 10.1186/s12864-020-6665-2. BMC Genomics. 2020. PMID: 32228448 Free PMC article.
-
Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner.Nat Microbiol. 2021 Aug;6(8):1043-1054. doi: 10.1038/s41564-021-00920-0. Epub 2021 Jul 5. Nat Microbiol. 2021. PMID: 34226711 Free PMC article.
-
Understanding the impact of antibiotic perturbation on the human microbiome.Genome Med. 2020 Sep 28;12(1):82. doi: 10.1186/s13073-020-00782-x. Genome Med. 2020. PMID: 32988391 Free PMC article. Review.
Cited by
-
Druggable proteins of Alistipes reveal promising antimicrobial targets against chronic intestinal inflammation.Front Pharmacol. 2025 Apr 24;16:1555062. doi: 10.3389/fphar.2025.1555062. eCollection 2025. Front Pharmacol. 2025. PMID: 40342998 Free PMC article.
-
'Smart', microbiome-sparing antibacterial therapy with a focus on the novel Lolamicin: an overview.Infection. 2025 Apr 12. doi: 10.1007/s15010-025-02538-4. Online ahead of print. Infection. 2025. PMID: 40220252 Review.
-
A Mechanistic Approach to Replacing Antibiotics with Natural Products in the Treatment of Bacterial Diarrhea.Biomolecules. 2025 Jul 18;15(7):1045. doi: 10.3390/biom15071045. Biomolecules. 2025. PMID: 40723916 Free PMC article. Review.
References
-
- Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. . (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

