A study of antibiotic resistance pattern of clinical bacterial pathogens isolated from patients in a tertiary care hospital
- PMID: 38694800
- PMCID: PMC11061477
- DOI: 10.3389/fmicb.2024.1383989
A study of antibiotic resistance pattern of clinical bacterial pathogens isolated from patients in a tertiary care hospital
Abstract
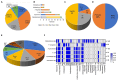
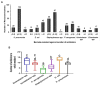
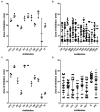
We investigated antibiotic resistance pattern in clinical bacterial pathogens isolated from in-patients and out-patients, and compared it with non-clinical bacterial isolates. 475 bacterial strains isolated from patients were examined for antibiotic resistance. Staphylococcus spp. (148; 31.1%) were found to be the most prevalent, followed by Klebsiella pneumoniae (135; 28.4%), Escherichia coli (74; 15.5%), Pseudomonas aeruginosa (65; 13.6%), Enterobacter spp. (28; 5.8%), and Acinetobacter spp. (25; 5.2%). Drug-resistant bacteria isolated were extended spectrum-β-lactamase K. pneumoniae (8.8%), E. coli (20%), metallo-β-lactamase P. aeruginosa (14; 2.9%), erythromycin-inducing clindamycin resistant (7.4%), and methicillin-resistant Staphylococcus species (21.6%). Pathogens belonging to the Enterobacteriaceae family were observed to undergo directional selection developing resistance against antibiotics ciprofloxacin, piperacillin-tazobactam, cefepime, and cefuroxime. Pathogens in the surgical ward exhibited higher levels of antibiotic resistance, while non-clinical P. aeruginosa and K. pneumoniae strains were more antibiotic-susceptible. Our research assisted in identifying the drugs that can be used to control infections caused by antimicrobial resistant bacteria in the population and in monitoring the prevalence of drug-resistant bacterial pathogens.
Keywords: antimicrobial resistance; erythromycin-induced clindamycin resistance; extended spectrum-β-lactamase; metallo-β-lactamase; methicillin-resistant Staphylococcus aureus.
Copyright © 2024 Handa, Patel, Bhattacharya, Kothari, Kavathia and Vyas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Akiba M., Senba H., Otagiri H., Prabhasankar V. P., Taniyasu S., Yamashita N., et al. (2015). Impact of wastewater from different sources on the prevalence of antimicrobial-resistant Escherichia coli in sewage treatment plants in South India. Ecotoxicol. Environ. Saf. 115, 203–208. doi: 10.1016/j.ecoenv.2015.02.018, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

