Population characteristics of pathogenic Escherichia coli in puerperal metritis of dairy cows in Ningxia region of China: a systemic taxa distribution of virulence factors and drug resistance genes
- PMID: 38694808
- PMCID: PMC11061491
- DOI: 10.3389/fmicb.2024.1364373
Population characteristics of pathogenic Escherichia coli in puerperal metritis of dairy cows in Ningxia region of China: a systemic taxa distribution of virulence factors and drug resistance genes
Abstract
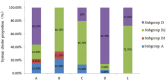
Escherichia coli (E. coli) is closely associated with the occurrence of puerperal metritis in dairy cows. E. coli carries some the virulence and multi-drug resistant genes, which pose a serious threat to the health of postpartum cows. In this study, E. coli was isolated and identified from the uterine contents of postpartum cows with puerperal metritis in the Ningxia region of China, and its phylogenetic subgroups were determined. Meanwhile, virulence and drug resistance genes carried by E. coli and drug sensitivity were detected, and the characteristics of virulence and drug resistance genes distribution in E. coli phylogroups were further analyzed. The results showed that the isolation rate of E. coli in puerperal metritis samples was 95.2%. E. coli was mainly divided into phylogroups B2 and D, followed by groups A and B1, and was more connected to O157:H7, O169:H4, and ECC-1470 type strains. The virulence genes were mainly dominated by ompF (100%), traT (100%), fimH (97%), papC (96%), csgA (95%), Ang43 (93.9%), and ompC (93%), and the resistance genes were dominated by TEM (99%), tetA (71.7%), aac(3)II (66.7%), and cmlA (53.5%). Additionally, it was observed that the virulence and resistance gene phenotypes could be divided into two subgroups, with subgroup B2 and D having the highest distributions. Drug sensitivity tests also revealed that the E. coli was most sensitive to the fluoroquinolones enrofloxacin, followed by macrolides, aminoglycosides, tetracyclines, β-lactams, peptides and sulfonamides, and least sensitive to lincosamides. These results imply that pathogenic E. coli, which induces puerperal metritis of dairy cows in the Ningxia region of China, primarily belongs to the group B2 and D, contains multiple virulence and drug resistance genes, Moreover, E. coli has evolved resistance to several drugs including penicillin, lincomycin, cotrimoxazole, and streptomycin. It will offer specific guidelines reference for the prevention and treatment of puerperal metritis in dairy cows with E. coli infections in the Ningxia region of China.
Keywords: Escherichia coli; dairy cows; drug resistance gene; drug sensitivity; puerperal metritis; virulence gene.
Copyright © 2024 Wei, Ding, Wang, Luo, Zhao and Dan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Abd E. R., Ibrahim R. A., Mohamed D. S., Ahmed E. F., Hashem Z. S. (2020). Prevalence of virulence genes and their association with antimicrobial resistance among pathogenic E. coli isolated from egyptian patients with different clinical infections. Infect Drug Resist. 13, 1221–1236. doi: 10.2147/IDR.S241073 - DOI - PMC - PubMed
-
- Aguirre-Sanchez J. R., Valdez-Torres J. B., Del C. N., Martinez-Urtaza J., Del C. N., Lee B. G., et al. (2022). Phylogenetic group and virulence profile classification in Escherichia coli from distinct isolation sources in mexico. Infect. Genet. Evol. 106:105380. doi: 10.1016/j.meegid.2022.105380, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

