The MexTAg collaborative cross: host genetics affects asbestos related disease latency, but has little influence once tumours develop
- PMID: 38694815
- PMCID: PMC11061428
- DOI: 10.3389/ftox.2024.1373003
The MexTAg collaborative cross: host genetics affects asbestos related disease latency, but has little influence once tumours develop
Abstract
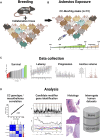
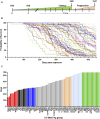
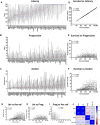
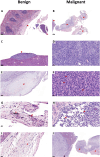
Objectives: This study combines two innovative mouse models in a major gene discovery project to assess the influence of host genetics on asbestos related disease (ARD). Conventional genetics studies provided evidence that some susceptibility to mesothelioma is genetic. However, the identification of host modifier genes, the roles they may play, and whether they contribute to disease susceptibility remain unknown. Here we report a study designed to rapidly identify genes associated with mesothelioma susceptibility by combining the Collaborative Cross (CC) resource with the well-characterised MexTAg mesothelioma mouse model. Methods: The CC is a powerful mouse resource that harnesses over 90% of common genetic variation in the mouse species, allowing rapid identification of genes mediating complex traits. MexTAg mice rapidly, uniformly, and predictably develop mesothelioma, but only after asbestos exposure. To assess the influence of host genetics on ARD, we crossed 72 genetically distinct CC mouse strains with MexTAg mice and exposed the resulting CC-MexTAg (CCMT) progeny to asbestos and monitored them for traits including overall survival, the time to ARD onset (latency), the time between ARD onset and euthanasia (disease progression) and ascites volume. We identified phenotype-specific modifier genes associated with these traits and we validated the role of human orthologues in asbestos-induced carcinogenesis using human mesothelioma datasets. Results: We generated 72 genetically distinct CCMT strains and exposed their progeny (2,562 in total) to asbestos. Reflecting the genetic diversity of the CC, there was considerable variation in overall survival and disease latency. Surprisingly, however, there was no variation in disease progression, demonstrating that host genetic factors do have a significant influence during disease latency but have a limited role once disease is established. Quantitative trait loci (QTL) affecting ARD survival/latency were identified on chromosomes 6, 12 and X. Of the 97-protein coding candidate modifier genes that spanned these QTL, eight genes (CPED1, ORS1, NDUFA1, HS1BP3, IL13RA1, LSM8, TES and TSPAN12) were found to significantly affect outcome in both CCMT and human mesothelioma datasets. Conclusion: Host genetic factors affect susceptibility to development of asbestos associated disease. However, following mesothelioma establishment, genetic variation in molecular or immunological mechanisms did not affect disease progression. Identification of multiple candidate modifier genes and their human homologues with known associations in other advanced stage or metastatic cancers highlights the complexity of ARD and may provide a pathway to identify novel therapeutic targets.
Keywords: MexTAg; asbestos related disease; collaborative cross; gene discovery; host genetics; mesothelioma; mouse model.
Copyright © 2024 Fisher, Patrick, Hoang, Marcq, Behrouzfar, Young, Miller, Robinson, Bueno, Nowak, Lesterhuis, Morahan and Lake.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Australia Co: Australian code for the care and use of animals for scientific purposes. In. Edited by N. H. M. R. Council, 8th edn. Canberra: Commonwaealth or Australia; 2013.
LinkOut - more resources
Full Text Sources

