Identification of immune-related biomarkers for glaucoma using gene expression profiling
- PMID: 38694874
- PMCID: PMC11062407
- DOI: 10.3389/fgene.2024.1366453
Identification of immune-related biomarkers for glaucoma using gene expression profiling
Abstract
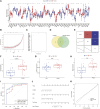
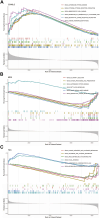
Introduction: Glaucoma, a principal cause of irreversible vision loss, is characterized by intricate optic neuropathy involving significant immune mechanisms. This study seeks to elucidate the molecular and immune complexities of glaucoma, aiming to improve our understanding of its pathogenesis. Methods: Gene expression profiles from glaucoma patients were analyzed to identify immune-related differentially expressed genes (DEGs). Techniques used were weighted gene co-expression network analysis (WGCNA) for network building, machine learning algorithms for biomarker identification, establishment of subclusters related to immune reactions, and single-sample gene set enrichment analysis (ssGSEA) to explore hub genes' relationships with immune cell infiltration and immune pathway activation. Validation was performed using an NMDA-induced excitotoxicity model and RT-qPCR for hub gene expression measurement. Results: The study identified 409 DEGs differentiating healthy individuals from glaucoma patients, highlighting the immune response's significance in disease progression. Immune cell infiltration analysis revealed elevated levels of activated dendritic cells, natural killer cells, monocytes, and immature dendritic cells in glaucoma samples. Three hub genes, CD40LG, TEK, and MDK, were validated as potential diagnostic biomarkers for high-risk glaucoma patients, showing increased expression in the NMDA-induced excitotoxicity model. Discussion: The findings propose the three identified immune-related genes (IRGs) as novel diagnostic markers for glaucoma, offering new insights into the disease's pathogenesis and potential therapeutic targets. The strong correlation between these IRGs and immune responses underscores the intricate role of immunity in glaucoma, suggesting a shift in the approach to its diagnosis and treatment.
Keywords: WGCNA; biomarkers; glaucoma; immune cell infiltration; immune-related genes; machine learning; subclusters.
Copyright © 2024 Wang, Pu, Tan, Wang, Zeng, Lei, Gao and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

