Monocyte Production of C1q Potentiates CD8+ T-Cell Function Following Respiratory Viral Infection
- PMID: 38696270
- PMCID: PMC11376238
- DOI: 10.1165/rcmb.2024-0004OC
Monocyte Production of C1q Potentiates CD8+ T-Cell Function Following Respiratory Viral Infection
Abstract
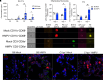
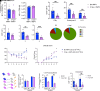
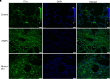
Respiratory viral infections remain a leading cause of morbidity and mortality. Using a murine model of human metapneumovirus, we identified recruitment of a C1q-expressing inflammatory monocyte population concomitant with viral clearance by adaptive immune cells. Genetic ablation of C1q led to reduced CD8+ T-cell function. Production of C1q by a myeloid lineage was necessary to enhance CD8+ T-cell function. Activated and dividing CD8+ T cells expressed a C1q receptor, gC1qR. Perturbation of gC1qR signaling led to altered CD8+ T-cell IFN-γ production, metabolic capacity, and cell proliferation. Autopsy specimens from fatal respiratory viral infections in children exhibited diffuse production of C1q by an interstitial population. Humans with severe coronavirus disease (COVID-19) infection also exhibited upregulation of gC1qR on activated and rapidly dividing CD8+ T cells. Collectively, these studies implicate C1q production from monocytes as a critical regulator of CD8+ T-cell function following respiratory viral infection.
Keywords: COVID-19; antiviral immunity; complement; human metapneumovirus.
Figures



Update of
-
Monocyte production of C1q potentiates CD8 + T cell effector function following respiratory viral infection.bioRxiv [Preprint]. 2023 Jun 6:2023.06.04.543430. doi: 10.1101/2023.06.04.543430. bioRxiv. 2023. Update in: Am J Respir Cell Mol Biol. 2024 Sep;71(3):294-306. doi: 10.1165/rcmb.2024-0004OC. PMID: 37333212 Free PMC article. Updated. Preprint.
Comment in
-
Monocytes: See One Queuing Local Adaptive Immune Responses to Respiratory Viruses.Am J Respir Cell Mol Biol. 2024 Sep;71(3):259-261. doi: 10.1165/rcmb.2024-0195ED. Am J Respir Cell Mol Biol. 2024. PMID: 38717817 Free PMC article. No abstract available.
References
-
- Li Y, Nair H. Trends in the global burden of lower respiratory infections: the knowns and the unknowns. Lancet Infect Dis . 2022;22:1523–1525. - PubMed
MeSH terms
Substances
Grants and funding
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- 75N93022D00005/AI/NIAID NIH HHS/United States
- F30 HL159915/HL/NHLBI NIH HHS/United States
- T32 AI138954/AI/NIAID NIH HHS/United States
- 75N93023D00005/AI/NIAID NIH HHS/United States
- K12 HD000850/HD/NICHD NIH HHS/United States
- T32 GM008208/GM/NIGMS NIH HHS/United States
- 75N99020D00005/OF/ORFDO NIH HHS/United States
- Henry L. Hillman Foundation/United States
- R01 AI085062/AI/NIAID NIH HHS/United States
- R01 HL137709/HL/NHLBI NIH HHS/United States
- T32GM008208/GM/NIGMS NIH HHS/United States
- 1F30HL159915/HL/NHLBI NIH HHS/United States
- 75N95020D00005/DA/NIDA NIH HHS/United States
- AI085062/National Institute of Allergy and Infectious Diseases/United States
- K12HD000850/Eunice Kennedy Shriver National Institute of Child Health and Human Development/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

