DELLA proteins recruit the Mediator complex subunit MED15 to coactivate transcription in land plants
- PMID: 38696472
- PMCID: PMC11087773
- DOI: 10.1073/pnas.2319163121
DELLA proteins recruit the Mediator complex subunit MED15 to coactivate transcription in land plants
Abstract
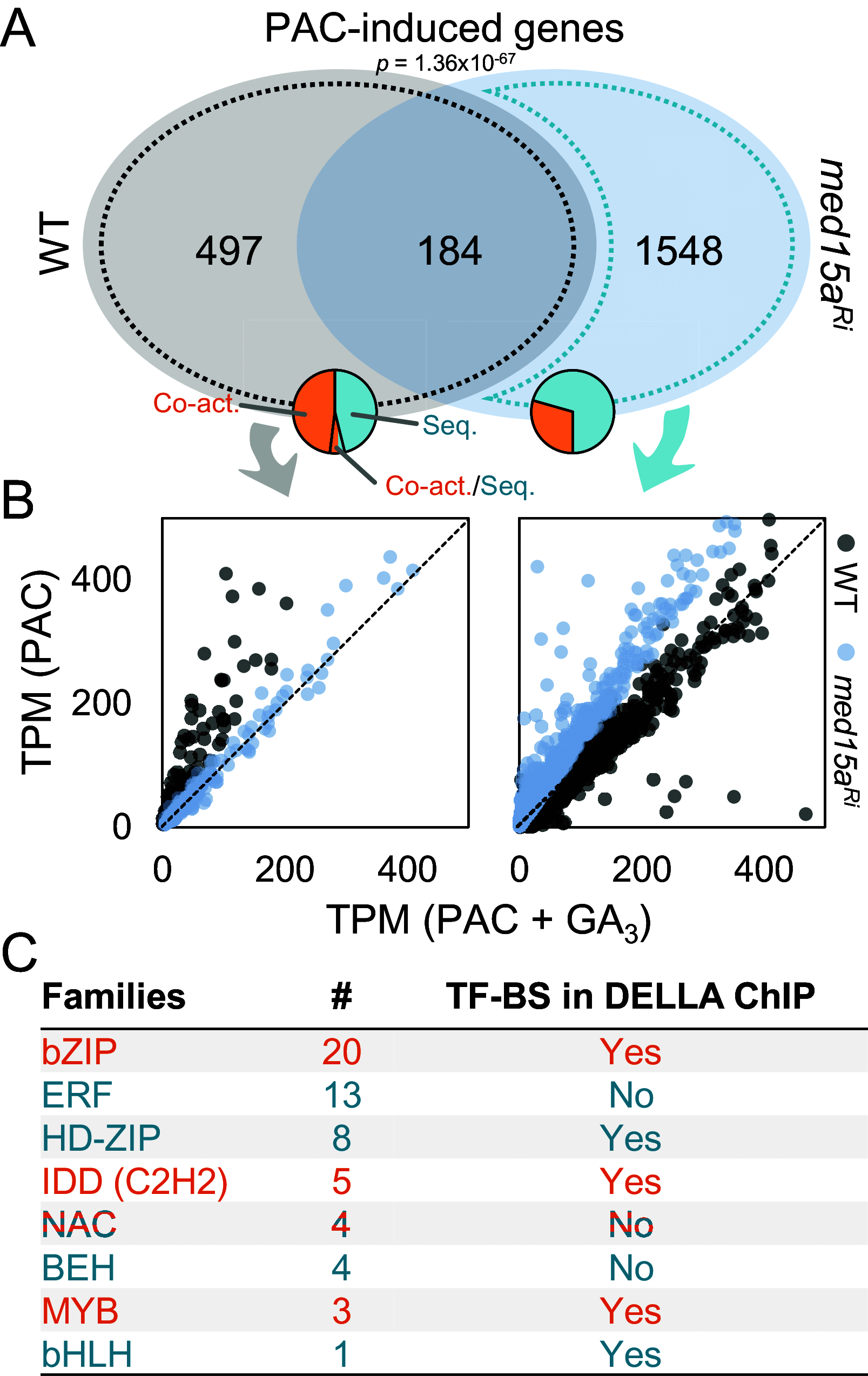
DELLA proteins are negative regulators of the gibberellin response pathway in angiosperms, acting as central hubs that interact with hundreds of transcription factors (TFs) and regulators to modulate their activities. While the mechanism of TF sequestration by DELLAs to prevent DNA binding to downstream targets has been extensively documented, the mechanism that allows them to act as coactivators remains to be understood. Here, we demonstrate that DELLAs directly recruit the Mediator complex to specific loci in Arabidopsis, facilitating transcription. This recruitment involves DELLA amino-terminal domain and the conserved MED15 KIX domain. Accordingly, partial loss of MED15 function mainly disrupted processes known to rely on DELLA coactivation capacity, including cytokinin-dependent regulation of meristem function and skotomorphogenic response, gibberellin metabolism feedback, and flavonol production. We have also found that the single DELLA protein in the liverwort Marchantia polymorpha is capable of recruiting MpMED15 subunits, contributing to transcriptional coactivation. The conservation of Mediator-dependent transcriptional coactivation by DELLA between Arabidopsis and Marchantia implies that this mechanism is intrinsic to the emergence of DELLA in the last common ancestor of land plants.
Keywords: DELLA proteins; Mediator; gibberellins; hormone signaling; transactivation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
-
- Hernández-García J., Briones-Moreno A., Blázquez M. A., Origin and evolution of gibberellin signaling and metabolism in plants. Semin. Cell Dev. Biol. 109, 46–54 (2021). - PubMed
-
- Sun T. P., The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21, R338–R345 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

