Collaborative Cross mice have diverse phenotypic responses to infection with Methicillin-resistant Staphylococcus aureus USA300
- PMID: 38696518
- PMCID: PMC11108197
- DOI: 10.1371/journal.pgen.1011229
Collaborative Cross mice have diverse phenotypic responses to infection with Methicillin-resistant Staphylococcus aureus USA300
Abstract
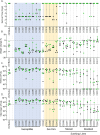
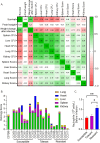
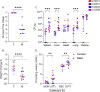
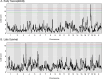
Staphylococcus aureus (S. aureus) is an opportunistic pathogen causing diseases ranging from mild skin infections to life threatening conditions, including endocarditis, pneumonia, and sepsis. To identify host genes modulating this host-pathogen interaction, we infected 25 Collaborative Cross (CC) mouse strains with methicillin-resistant S. aureus (MRSA) and monitored disease progression for seven days using a surgically implanted telemetry system. CC strains varied widely in their response to intravenous MRSA infection. We identified eight 'susceptible' CC strains with high bacterial load, tissue damage, and reduced survival. Among the surviving strains, six with minimal colonization were classified as 'resistant', while the remaining six tolerated higher organ colonization ('tolerant'). The kidney was the most heavily colonized organ, but liver, spleen and lung colonization were better correlated with reduced survival. Resistant strains had higher pre-infection circulating neutrophils and lower post-infection tissue damage compared to susceptible and tolerant strains. We identified four CC strains with sexual dimorphism: all females survived the study period while all males met our euthanasia criteria earlier. In these CC strains, males had more baseline circulating monocytes and red blood cells. We identified several CC strains that may be useful as new models for endocarditis, myocarditis, pneumonia, and resistance to MRSA infection. Quantitative Trait Locus (QTL) analysis identified two significant loci, on Chromosomes 18 and 3, involved in early susceptibility and late survival after infection. We prioritized Npc1 and Ifi44l genes as the strongest candidates influencing survival using variant analysis and mRNA expression data from kidneys within these intervals.
Copyright: © 2024 Nagarajan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

