A Workflow for Improved Analysis of Cross-Linking Mass Spectrometry Data Integrating Parallel Accumulation-Serial Fragmentation with MeroX and Skyline
- PMID: 38696819
- PMCID: PMC11099889
- DOI: 10.1021/acs.analchem.4c00829
A Workflow for Improved Analysis of Cross-Linking Mass Spectrometry Data Integrating Parallel Accumulation-Serial Fragmentation with MeroX and Skyline
Abstract
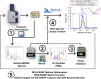
Cross-linking mass spectrometry (XL-MS) has evolved into a pivotal technique for probing protein interactions. This study describes the implementation of Parallel Accumulation-Serial Fragmentation (PASEF) on timsTOF instruments, enhancing the detection and analysis of protein interactions by XL-MS. Addressing the challenges in XL-MS, such as the interpretation of complex spectra, low abundant cross-linked peptides, and a data acquisition bias, our current study integrates a peptide-centric approach for the analysis of XL-MS data and presents the foundation for integrating data-independent acquisition (DIA) in XL-MS with a vendor-neutral and open-source platform. A novel workflow is described for processing data-dependent acquisition (DDA) of PASEF-derived information. For this, software by Bruker Daltonics is used, enabling the conversion of these data into a format that is compatible with MeroX and Skyline software tools. Our approach significantly improves the identification of cross-linked products from complex mixtures, allowing the XL-MS community to overcome current analytical limitations.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Niemeyer M.; Moreno Castillo E.; Ihling C. H.; Iacobucci C.; Wilde V.; Hellmuth A.; Hoehenwarter W.; Samodelov S. L.; Zurbriggen M. D.; Kastritis P. L.; Sinz A.; Calderón Villalobos L. I. A. Flexibility of Intrinsically Disordered Degrons in AUX/IAA Proteins Reinforces Auxin Co-Receptor Assemblies. Nat. Commun. 2020, 11 (1), 2277.10.1038/s41467-020-16147-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

