SAH is a major metabolic sensor mediating worsening metabolic crosstalk in metabolic syndrome
- PMID: 38696898
- PMCID: PMC11070633
- DOI: 10.1016/j.redox.2024.103139
SAH is a major metabolic sensor mediating worsening metabolic crosstalk in metabolic syndrome
Abstract
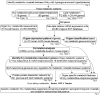
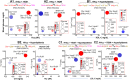
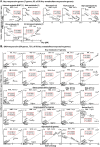
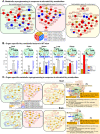
In this study, we observed worsening metabolic crosstalk in mouse models with concomitant metabolic disorders such as hyperhomocysteinemia (HHcy), hyperlipidemia, and hyperglycemia and in human coronary artery disease by analyzing metabolic profiles. We found that HHcy worsening is most sensitive to other metabolic disorders. To identify metabolic genes and metabolites responsible for the worsening metabolic crosstalk, we examined mRNA levels of 324 metabolic genes in Hcy, glucose-related and lipid metabolic systems. We examined Hcy-metabolites (Hcy, SAH and SAM) by LS-ESI-MS/MS in 6 organs (heart, liver, brain, lung, spleen, and kidney) from C57BL/6J mice. Through linear regression analysis of Hcy-metabolites and metabolic gene mRNA levels, we discovered that SAH-responsive genes were responsible for most metabolic changes and all metabolic crosstalk mediated by Serine, Taurine, and G3P. SAH-responsive genes worsen glucose metabolism and cause upper glycolysis activation and lower glycolysis suppression, indicative of the accumulation of glucose/glycogen and G3P, Serine synthesis inhibition, and ATP depletion. Insufficient Serine due to negative correlation of PHGDH with SAH concentration may inhibit the folate cycle and transsulfurarion pathway and consequential reduced antioxidant power, including glutathione, taurine, NADPH, and NAD+. Additionally, we identified SAH-activated pathological TG loop as the consequence of increased fatty acid (FA) uptake, FA β-oxidation and Ac-CoA production along with lysosomal damage. We concluded that HHcy is most responsive to other metabolic changes in concomitant metabolic disorders and mediates worsening metabolic crosstalk mainly via SAH-responsive genes, that organ-specific Hcy metabolism determines organ-specific worsening metabolic reprogramming, and that SAH, acetyl-CoA, Serine and Taurine are critical metabolites mediating worsening metabolic crosstalk, redox disturbance, hypomethylation and hyperacetylation linking worsening metabolic reprogramming in metabolic syndrome.
Keywords: Hyperhomocysteinemia; Metabolic syndrome; Redox; S-Adenosyl-homocysteine (SAH); Serine; Taurine.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no conflict of interest.
Figures







Similar articles
-
Aldh1l2 knockout mouse metabolomics links the loss of the mitochondrial folate enzyme to deregulation of a lipid metabolism observed in rare human disorder.Hum Genomics. 2020 Nov 9;14(1):41. doi: 10.1186/s40246-020-00291-3. Hum Genomics. 2020. PMID: 33168096 Free PMC article.
-
Examination of Genetic and Epigenetic Characteristics of Patients with Hyperhomocysteinemia Following High-Dose Folic Acid Consumption.Nutrients. 2025 Jun 27;17(13):2133. doi: 10.3390/nu17132133. Nutrients. 2025. PMID: 40647238 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
Cited by
-
The Roles of White Adipose Tissue and Liver NADPH in Dietary Restriction-Induced Longevity.Antioxidants (Basel). 2024 Jul 8;13(7):820. doi: 10.3390/antiox13070820. Antioxidants (Basel). 2024. PMID: 39061889 Free PMC article. Review.
-
AIMP3 maintains cardiac homeostasis by regulating the editing activity of methionyl-tRNA synthetase.Nat Cardiovasc Res. 2025 Jul;4(7):876-890. doi: 10.1038/s44161-025-00670-w. Epub 2025 Jun 25. Nat Cardiovasc Res. 2025. PMID: 40562875 Free PMC article.
-
Taurine alleviated paraquat-induced oxidative stress and gut-liver axis damage in weaned piglets by regulating the Nrf2/Keap1 and TLR4/NF-κB signaling pathways.J Anim Sci Biotechnol. 2025 Aug 18;16(1):117. doi: 10.1186/s40104-025-01244-3. J Anim Sci Biotechnol. 2025. PMID: 40826133 Free PMC article.
-
Brain Glycogen-Its Metabolic Role in Neuronal Health and Neurological Disorders-An Extensive Narrative Review.Metabolites. 2025 Feb 13;15(2):128. doi: 10.3390/metabo15020128. Metabolites. 2025. PMID: 39997753 Free PMC article. Review.
-
Cerebrospinal fluid analysis and changes over time in patients with subarachnoid hemorrhage: a prospective observational study.J Anesth Analg Crit Care. 2025 Jun 12;5(1):31. doi: 10.1186/s44158-025-00250-1. J Anesth Analg Crit Care. 2025. PMID: 40506779 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

