NRDE2 deficiency impairs homologous recombination repair and sensitizes hepatocellular carcinoma to PARP inhibitors
- PMID: 38697125
- PMCID: PMC11099347
- DOI: 10.1016/j.xgen.2024.100550
NRDE2 deficiency impairs homologous recombination repair and sensitizes hepatocellular carcinoma to PARP inhibitors
Abstract
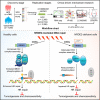
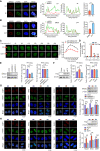
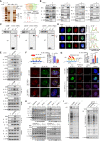
To identify novel susceptibility genes for hepatocellular carcinoma (HCC), we performed a rare-variant association study in Chinese populations consisting of 2,750 cases and 4,153 controls. We identified four HCC-associated genes, including NRDE2, RANBP17, RTEL1, and STEAP3. Using NRDE2 (index rs199890497 [p.N377I], p = 1.19 × 10-9) as an exemplary candidate, we demonstrated that it promotes homologous recombination (HR) repair and suppresses HCC. Mechanistically, NRDE2 binds to the subunits of casein kinase 2 (CK2) and facilitates the assembly and activity of the CK2 holoenzyme. This NRDE2-mediated enhancement of CK2 activity increases the phosphorylation of MDC1 and then facilitates the HR repair. These functions are eliminated almost completely by the NRDE2-p.N377I variant, which sensitizes the HCC cells to poly(ADP-ribose) polymerase (PARP) inhibitors, especially when combined with chemotherapy. Collectively, our findings highlight the relevance of the rare variants to genetic susceptibility to HCC, which would be helpful for the precise treatment of this malignancy.
Keywords: NRDE2; PARP inhibitors; hepatocellular carcinoma; homologous recombination repair; rare variant.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. C.A. Cancer J. Clin. 2019;69:7–34. - PubMed
-
- Cao P., Yang A., Wang R., Xia X., Zhai Y., Li Y., Yang F., Cui Y., Xie W., Liu Y., et al. Germline Duplication of SNORA18L5 Increases Risk for HBV-related Hepatocellular Carcinoma by Altering Localization of Ribosomal Proteins and Decreasing Levels of p53. Gastroenterology. 2018;155:542–556. - PubMed
-
- Kumar V., Kato N., Urabe Y., Takahashi A., Muroyama R., Hosono N., Otsuka M., Tateishi R., Omata M., Nakagawa H., et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat. Genet. 2011;43:455–458. - PubMed
-
- Li S., Fell S.M., Surova O., Smedler E., Wallis K., Chen Z.X., Hellman U., Johnsen J.I., Martinsson T., Kenchappa R.S., et al. The 1p36 Tumor Suppressor KIF 1Bbeta Is Required for Calcineurin Activation, Controlling Mitochondrial Fission and Apoptosis. Dev. Cell. 2016;36:164–178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

