RASSF4 Attenuates Metabolic Dysfunction-Associated Steatotic Liver Disease Progression via Hippo Signaling and Suppresses Hepatocarcinogenesis
- PMID: 38697356
- PMCID: PMC11217689
- DOI: 10.1016/j.jcmgh.2024.04.005
RASSF4 Attenuates Metabolic Dysfunction-Associated Steatotic Liver Disease Progression via Hippo Signaling and Suppresses Hepatocarcinogenesis
Abstract
Background & aims: Metabolic dysfunction-associated steatotic liver disease (MASLD) is a dynamic chronic liver disease closely related to metabolic abnormalities such as diabetes and obesity. MASLD can further progress to metabolic dysfunction-associated steatohepatitis (MASH), fibrosis, cirrhosis, and even hepatocellular carcinoma (HCC). However, the mechanisms underlying the progression of MASLD and further progression to liver fibrosis and liver cancer are unknown.
Methods: In this study, we performed transcriptome analysis in livers from mice with MASLD and found suppression of a potential anti-oncogene, RAS association domain protein 4 (RASSF4). RASSF4 expression levels were measured in liver or tumor tissues of patients with MASH or HCC, respectively. We established RASSF4 overexpression and knockout mouse models. The effects of RASSF4 were evaluated by quantitative polymerase chain reaction, Western blotting, histopathological analysis, wound healing assays, Transwell assays, EdU incorporation assays, colony formation assays, sorafenib sensitivity assays, and tumorigenesis assays.
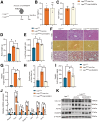
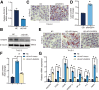
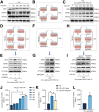
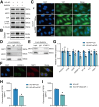
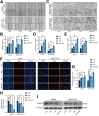
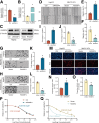
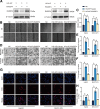
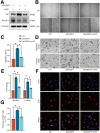
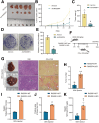
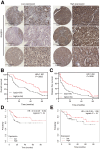
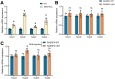
Results: RASSF4 was significantly down-regulated in MASH and HCC samples. Using liver-specific RASSF4 knockout mice, we demonstrated that loss of hepatic RASSF4 exacerbated hepatic steatosis and fibrosis. In contrast, RASSF4 overexpression prevented steatosis in MASLD mice. In addition, RASSF4 in hepatocytes suppressed the activation of hepatic stellate cells (HSCs) by reducing transforming growth factor beta secretion. Moreover, we found that RASSF4 is an independent prognostic factor for HCC. Mechanistically, we found that RASSF4 in the liver interacts with MST1 to inhibit YAP nuclear translocation through the Hippo pathway.
Conclusions: These findings establish RASSF4 as a therapeutic target for MASLD and HCC.
Keywords: HCC; Hepatic Stellate Cells; Hippo Pathway; MASLD; RASSF4.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Li J., Zou B., Yeo Y.H., Feng Y., et al. Prevalence, incidence, and outcome of non-alcoholic fatty liver disease in Asia, 1999-2019: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2019;4:389–398. - PubMed
-
- Zhou F., Zhou J., Wang W., et al. Unexpected rapid increase in the burden of NAFLD in China from 2008 to 2018: a systematic review and meta-analysis. Hepatology. 2019;70:1119–1133. - PubMed
-
- Ioannou G.N. Epidemiology and risk-stratification of NAFLD-associated HCC. J Hepatol. 2021;75:1476–1484. - PubMed
-
- El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

