Patient-derived follicular lymphoma spheroids recapitulate lymph node signaling and immune profile uncovering galectin-9 as a novel immunotherapeutic target
- PMID: 38697976
- PMCID: PMC11636880
- DOI: 10.1038/s41408-024-01041-7
Patient-derived follicular lymphoma spheroids recapitulate lymph node signaling and immune profile uncovering galectin-9 as a novel immunotherapeutic target
Abstract
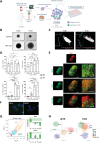
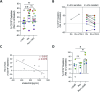
Follicular lymphoma (FL), the most common indolent non-Hodgkin lymphoma, constitutes a paradigm of immune tumor microenvironment (TME) contribution to disease onset, progression, and heterogenous clinical outcome. Here we present the first FL-Patient Derived Lymphoma Spheroid (FL-PDLS), including fundamental immune actors and features of TME in FL lymph nodes (LNs). FL-PDLS is organized in disc-shaped 3D structures composed of proliferating B and T cells, together with macrophages with an intermediate M1/M2 phenotype. FL-PDLS recapitulates the most relevant B-cell transcriptional pathways present in FL-LN (proliferation, epigenetic regulation, mTOR, adaptive immune system, among others). The T cell compartment in the FL-PDLS preserves CD4 subsets (follicular helper, regulatory, and follicular regulatory), also encompassing the spectrum of activation/exhaustion phenotypes in CD4 and CD8 populations. Moreover, this system is suitable for chemo and immunotherapy testing, recapitulating results obtained in the clinic. FL-PDLS allowed uncovering that soluble galectin-9 limits rituximab, rituximab, plus nivolumab/TIM-3 antitumoral activities. Blocking galectin-9 improves rituximab efficacy, highlighting galectin-9 as a novel immunotherapeutic target in FL. In conclusion, FL-PDLS maintains the crosstalk between malignant B cells and the immune LN-TME and constitutes a robust and multiplexed pre-clinical tool to perform drug screening in a patient-derived system, advancing toward personalized therapeutic approaches.
© 2024. The Author(s).
Conflict of interest statement
MN, RM, PB-L, and J-ML are employees of Imactiv3D. The rest of the authors declare no conflict of interest.
Figures



References
-
- Link BK, Maurer MJ, Nowakowski GS, Ansell SM, MacOn WR, Syrbu SI, et al. Rates and outcomes of follicular lymphoma transformation in the immunochemotherapy era: a report from the University of Iowa/Mayo Clinic specialized program of research excellence molecular epidemiology resource. J Clin Oncol. 2013;31:3272–8. - PMC - PubMed
-
- Jacobsen E. Follicular lymphoma: 2023 update on diagnosis and management. Am J Hematol. 2022;97:1638–51. - PubMed
-
- Carbone A, Roulland S, Gloghini A, Younes A, von Keudell G, López-Guillermo A, et al. Follicular lymphoma. Nat Rev Dis Prim. 2019;5:83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- IMLINFO EFA281/16/EC | European Regional Development Fund (Europski Fond za Regionalni Razvoj)
- TAIFOL 201933-30/Fundació la Marató de TV3 (TV3 Marathon Foundation)
- 2021SGR01294/Generalitat de Catalunya (Government of Catalonia)
- SAF2017/88275R/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- PRE2018-083797/Ministerio de Economía y Competitividad (Ministry of Economy and Competitiveness)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

