Globally occurring pelagiphage infections create ribosome-deprived cells
- PMID: 38698041
- PMCID: PMC11066056
- DOI: 10.1038/s41467-024-48172-w
Globally occurring pelagiphage infections create ribosome-deprived cells
Abstract
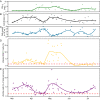
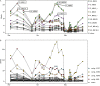
Phages play an essential role in controlling bacterial populations. Those infecting Pelagibacterales (SAR11), the dominant bacteria in surface oceans, have been studied in silico and by cultivation attempts. However, little is known about the quantity of phage-infected cells in the environment. Using fluorescence in situ hybridization techniques, we here show pelagiphage-infected SAR11 cells across multiple global ecosystems and present evidence for tight community control of pelagiphages on the SAR11 hosts in a case study. Up to 19% of SAR11 cells were phage-infected during a phytoplankton bloom, coinciding with a ~90% reduction in SAR11 cell abundance within 5 days. Frequently, a fraction of the infected SAR11 cells were devoid of detectable ribosomes, which appear to be a yet undescribed possible stage during pelagiphage infection. We dubbed such cells zombies and propose, among other possible explanations, a mechanism in which ribosomal RNA is used as a resource for the synthesis of new phage genomes. On a global scale, we detected phage-infected SAR11 and zombie cells in the Atlantic, Pacific, and Southern Oceans. Our findings illuminate the important impact of pelagiphages on SAR11 populations and unveil the presence of ribosome-deprived zombie cells as part of the infection cycle.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
- AWI_PS133/1_06/Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research (Alfred-Wegener- Institute, Helmholtz Centre for Polar and Marine Research)
- 03G0245A/Bundesministerium für Bildung, Wissenschaft und Kultur (Federal Ministry of Education, Science and Culture)
- 42076105/National Natural Science Foundation of China (National Science Foundation of China)
- FOR 2406/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources

