H3K27me3 timely dictates uterine epithelial transcriptome remodeling and thus transformation essential for normal embryo implantation
- PMID: 38698061
- PMCID: PMC11303564
- DOI: 10.1038/s41418-024-01302-9
H3K27me3 timely dictates uterine epithelial transcriptome remodeling and thus transformation essential for normal embryo implantation
Abstract
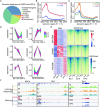
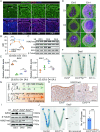
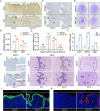
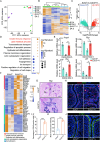
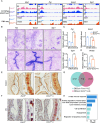
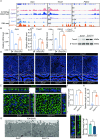
Uterine luminal epithelia (LE), the first layer contacting with the blastocyst, acquire receptivity for normal embryo implantation. Besides the well-accepted transcriptional regulation dominated by ovarian estrogen and progesterone for receptivity establishment, the involvement of epigenetic mechanisms remains elusive. This study systematically profiles the transcriptome and genome-wide H3K27me3 distribution in the LE throughout the preimplantation. Combining genetic and pharmacological approaches targeting the PRC2 core enzyme Ezh1/2, we demonstrate that the defective remodeling of H3K27me3 in the preimplantation stage disrupts the differentiation of LE, and derails uterine receptivity, resulting in implantation failure. Specifically, crucial epithelial genes, Pgr, Gata2, and Sgk1, are transcriptionally silenced through de novo deposition of H3K27me3 for LE transformation, and their sustained expression in the absence of H3K27me3 synergistically confines the nuclear translocation of FOXO1. Further functional studies identify several actin-associated genes, including Arpin, Tmod1, and Pdlim2, as novel direct targets of H3K27me3. Their aberrantly elevated expression impedes the morphological remodeling of LE, a hindrance alleviated by treatment with cytochalasin D which depolymerizes F-actin. Collectively, this study uncovers a previously unappreciated epigenetic regulatory mechanism for the transcriptional silencing of key LE genes via H3K27me3, essential for LE differentiation and thus embryo implantation.
© 2024. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Understanding epigenetic regulation in the endometrium - lessons from mouse models with implantation defects.Epigenomics. 2025 Jun;17(8):541-554. doi: 10.1080/17501911.2025.2491298. Epub 2025 Apr 14. Epigenomics. 2025. PMID: 40228031 Review.
-
Podocalyxin is a key negative regulator of human endometrial epithelial receptivity for embryo implantation.Hum Reprod. 2021 Apr 20;36(5):1353-1366. doi: 10.1093/humrep/deab032. Hum Reprod. 2021. PMID: 33822049 Free PMC article.
-
Arsenic disrupts H3K9me3 and H3K27me3 balance by biasing PRC2.1 and PRC2.2 activity via PALI1 inhibition in carcinogenesis.Int J Biol Sci. 2025 Jun 9;21(9):4069-4080. doi: 10.7150/ijbs.115605. eCollection 2025. Int J Biol Sci. 2025. PMID: 40612682 Free PMC article.
-
WT1 directs normal progesterone receptor-chromatin binding essential for uterine receptivity at peri-implantation.Proc Natl Acad Sci U S A. 2025 Jul 15;122(28):e2504361122. doi: 10.1073/pnas.2504361122. Epub 2025 Jul 8. Proc Natl Acad Sci U S A. 2025. PMID: 40627402
-
Clinical outcomes following selection of human preimplantation embryos with time-lapse monitoring: a systematic review.Hum Reprod Update. 2014 Sep-Oct;20(5):617-31. doi: 10.1093/humupd/dmu023. Epub 2014 Jun 2. Hum Reprod Update. 2014. PMID: 24890606
Cited by
-
Decoding Müllerian Duct Epithelial Regionalization.Mol Reprod Dev. 2025 Feb;92(2):e70018. doi: 10.1002/mrd.70018. Mol Reprod Dev. 2025. PMID: 39994938 Free PMC article. Review.
-
Understanding epigenetic regulation in the endometrium - lessons from mouse models with implantation defects.Epigenomics. 2025 Jun;17(8):541-554. doi: 10.1080/17501911.2025.2491298. Epub 2025 Apr 14. Epigenomics. 2025. PMID: 40228031 Review.
-
Endometrial aging is accompanied by H3K27ac and PGR loss.Nat Aging. 2025 May;5(5):816-830. doi: 10.1038/s43587-025-00859-5. Epub 2025 May 20. Nat Aging. 2025. PMID: 40394215 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

