Oral bacteria relative abundance in faeces increases due to gut microbiota depletion and is linked with patient outcomes
- PMID: 38698178
- PMCID: PMC11152985
- DOI: 10.1038/s41564-024-01680-3
Oral bacteria relative abundance in faeces increases due to gut microbiota depletion and is linked with patient outcomes
Abstract
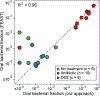
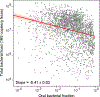
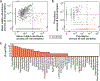
The detection of oral bacteria in faecal samples has been associated with inflammation and intestinal diseases. The increased relative abundance of oral bacteria in faeces has two competing explanations: either oral bacteria invade the gut ecosystem and expand (the 'expansion' hypothesis), or oral bacteria transit through the gut and their relative increase marks the depletion of other gut bacteria (the 'marker' hypothesis). Here we collected oral and faecal samples from mouse models of gut dysbiosis (antibiotic treatment and DSS-induced colitis) and used 16S ribosomal RNA sequencing to determine the abundance dynamics of oral bacteria. We found that the relative, but not absolute, abundance of oral bacteria increases, reflecting the 'marker' hypothesis. Faecal microbiome datasets from diverse patient cohorts, including healthy individuals and patients with allogeneic haematopoietic cell transplantation or inflammatory bowel disease, consistently support the 'marker' hypothesis and explain associations between oral bacterial abundance and patient outcomes consistent with depleted gut microbiota. By distinguishing between the two hypotheses, our study guides the interpretation of microbiome compositional data and could potentially identify cases where therapies are needed to rebuild the resident microbiome rather than protect against invading oral bacteria.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
J.U.P. reports research funding, intellectual property fees and travel reimbursement from Seres Therapeutics and consulting fees from DaVolterra, CSL Behring and from Maat Pharma. He serves on an Advisory board of and holds equity in Postbiotics Plus Research. He has filed intellectual property applications related to the microbiome (reference nos. 62/843,849, 62/977,908 and 15/756,845). M.R.M.v.d.B. has received research support from Seres Therapeutics; he has consulted, received honorarium from or participated in advisory boards for Seres Therapeutics, WindMIL Therapeutics, Rheos, Frazier Healthcare Partners, Nektar Therapeutics, Notch Therapeutics, Forty Seven Inc., Priothera, Ceramedix, Lygenesis, Pluto Immunotherapeutics, Magenta Therapeutics, Merck & Co., Inc. and DKMS Medical Council (Board); and he has IP Licensing with Seres Therapeutics, Juno Therapeutics and stock options from Seres and Notch Therapeutics. T.M.H. has participated in a scientific advisory board for Boehringer-Ingelheim Inc. T.R. is currently an employee of BioNTech SE. Memorial Sloan Kettering Cancer Center (MSKCC) has financial interests relative to Seres Therapeutics. The remaining authors declare no competing interests.
Figures




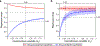
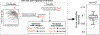

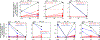

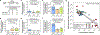


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

