CXCR6-positive circulating mucosal-associated invariant T cells can identify patients with non-small cell lung cancer responding to anti-PD-1 immunotherapy
- PMID: 38698468
- PMCID: PMC11067263
- DOI: 10.1186/s13046-024-03046-3
CXCR6-positive circulating mucosal-associated invariant T cells can identify patients with non-small cell lung cancer responding to anti-PD-1 immunotherapy
Abstract
Background: Mucosal-associated invariant T (MAIT) cells have been reported to regulate tumor immunity. However, the immune characteristics of MAIT cells in non-small cell lung cancer (NSCLC) and their correlation with the treatment efficacy of immune checkpoint inhibitors (ICIs) remain unclear.
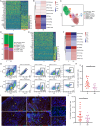
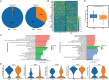
Patients and methods: In this study, we performed single-cell RNA sequencing (scRNA-seq), flow cytometry, and multiplex immunofluorescence assays to determine the proportion and characteristics of CD8+MAIT cells in patients with metastatic NSCLC who did and did not respond to anti-PD-1 therapy. Survival analyses were employed to determine the effects of MAIT proportion and C-X-C chemokine receptor 6 (CXCR6) expression on the prognosis of patients with advanced NSCLC.
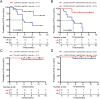
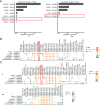
Results: The proportion of activated and proliferating CD8+MAIT cells were significantly higher in responders-derived peripheral blood mononuclear cells (PBMCs) and lung tissues before anti-PD-1 therapy, with enhanced expression of cytotoxicity-related genes including CCL4, KLRG1, PRF1, NCR3, NKG7, GZMB, and KLRK1. The responders' peripheral and tumor-infiltrating CD8+MAIT cells showed an upregulated CXCR6 expression. Similarly, CXCR6+CD8+MAIT cells from responders showed higher expression of cytotoxicity-related genes, such as CST7, GNLY, KLRG1, NKG7, and PRF1. Patients with ≥15.1% CD8+MAIT cells to CD8+T cells ratio and ≥35.9% CXCR6+CD8+MAIT cells to CD8+MAIT cells ratio in peripheral blood showed better progression-free survival (PFS) after immunotherapy. The role of CD8+MAIT cells in lung cancer immunotherapy was potentially mediated by classical/non-classical monocytes through the CXCL16-CXCR6 axis.
Conclusion: CD8+MAIT cells are a potential predictive biomarker for patients with NSCLC responding to anti-PD-1 therapy. The correlation between CD8+MAIT cells and immunotherapy sensitivity may be ascribed to high CXCR6 expression.
Keywords: CXCR6; Circulating mucosal-associated invariant T cells; Immunotherapy; Non-small cell lung cancer; Single-cell RNA-sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. The 2015 World Health Organization Classification of Lung Tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–60. doi: 10.1097/JTO.0000000000000630. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

