The prognostic and immunological role of MCM3 in pan-cancer and validation of prognosis in a clinical lower-grade glioma cohort
- PMID: 38698811
- PMCID: PMC11063780
- DOI: 10.3389/fphar.2024.1390615
The prognostic and immunological role of MCM3 in pan-cancer and validation of prognosis in a clinical lower-grade glioma cohort
Abstract
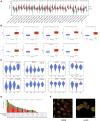
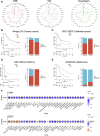
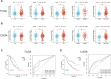
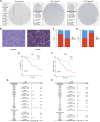
Background: Previous studies have shown that MCM3 plays a key role in initiating DNA replication. However, the mechanism of MCM3 function in most cancers is still unknown. The aim of our study was to explore the expression, prognostic role, and immunological characteristics of MCM3 across cancers. Methods: We explored the expression pattern of MCM3 across cancers. We subsequently explored the prognostic value of MCM3 expression by using univariate Cox regression analysis. Spearman correlation analysis was performed to determine the correlations between MCM3 and immune-related characteristics, mismatching repair (MMR) signatures, RNA modulator genes, cancer stemness, programmed cell death (PCD) gene expression, tumour mutation burden (TMB), microsatellite instability (MSI), and neoantigen levels. The role of MCM3 in predicting the response to immune checkpoint blockade (ICB) therapy was further evaluated in four immunotherapy cohorts. Single-cell data from CancerSEA were analysed to assess the biological functions associated with MCM3 in 14 cancers. The clinical correlation and independent prognostic significance of MCM3 were further analysed in the TCGA and CGGA lower-grade glioma (LGG) cohorts, and a prognostic nomogram was constructed. Immunohistochemistry in a clinical cohort was utilized to validate the prognostic utility of MCM3 expression in LGG. Results: MCM3 expression was upregulated in most tumours and strongly associated with patient outcomes in many cancers. Correlation analyses demonstrated that MCM3 expression was closely linked to immune cell infiltration, immune checkpoints, MMR genes, RNA modulator genes, cancer stemness, PCD genes and the TMB in most tumours. There was an obvious difference in outcomes between patients with high MCM3 expression and those with low MCM3 expression in the 4 ICB treatment cohorts. Single-cell analysis indicated that MCM3 was mainly linked to the cell cycle, DNA damage and DNA repair. The expression of MCM3 was associated with the clinical features of LGG patients and was an independent prognostic indicator. Finally, the prognostic significance of MCM3 in LGG was validated in a clinical cohort. Conclusion: Our study suggested that MCM3 can be used as a potential prognostic marker for cancers and may be associated with tumour immunity. In addition, MCM3 is a promising predictor of immunotherapy responses.
Keywords: immunotherapy; lower-grade glioma; minichromosome maintenance complex component 3 (MCM3); pan-cancer; prognosis.
Copyright © 2024 Huang, Jiang, Tan, Nong, Yan, Yang, Mo, Ling, Deng and Gong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

