Virtual brain twins: from basic neuroscience to clinical use
- PMID: 38698901
- PMCID: PMC11065363
- DOI: 10.1093/nsr/nwae079
Virtual brain twins: from basic neuroscience to clinical use
Abstract
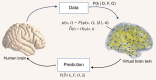
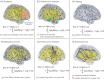
Virtual brain twins are personalized, generative and adaptive brain models based on data from an individual's brain for scientific and clinical use. After a description of the key elements of virtual brain twins, we present the standard model for personalized whole-brain network models. The personalization is accomplished using a subject's brain imaging data by three means: (1) assemble cortical and subcortical areas in the subject-specific brain space; (2) directly map connectivity into the brain models, which can be generalized to other parameters; and (3) estimate relevant parameters through model inversion, typically using probabilistic machine learning. We present the use of personalized whole-brain network models in healthy ageing and five clinical diseases: epilepsy, Alzheimer's disease, multiple sclerosis, Parkinson's disease and psychiatric disorders. Specifically, we introduce spatial masks for relevant parameters and demonstrate their use based on the physiological and pathophysiological hypotheses. Finally, we pinpoint the key challenges and future directions.
Keywords: brain disorder; inference; neuroscience; personalized modeling; virtual brain twin.
© The Author(s) 2024. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures

References
-
- Grieves MW. Virtually intelligent product systems: digital and physical twins. In: Flumerfelt S, Schwartz KG, Mavris D et al. (eds) Complex Systems Engineering: Theory and Practice. Reston, VA: American Institute of Aeronautics and Astronautics, 2019, 175–200.10.2514/5.9781624105654.0175.0200 - DOI
-
- Amunts K, Axer M, Bitsch L et al. The coming decade of digital brain research - a vision for neuroscience at the intersection of technology and computing (version 2.0). Zenodo; 2022, doi: 10.5281/zenodo.6630232.
-
- Kapteyn MG, Knezevic DJ, Huynh DB et al. Data-driven physics-based digital twins via a library of component-based reduced-order models. Int J Numer Meth Eng 2022; 123: 2986–3003.10.1002/nme.6423 - DOI
Publication types
LinkOut - more resources
Full Text Sources
