Post-transcriptional Regulation of Gene Expression via Unproductive Splicing
- PMID: 38698955
- PMCID: PMC11062102
- DOI: 10.32607/actanaturae.27337
Post-transcriptional Regulation of Gene Expression via Unproductive Splicing
Abstract
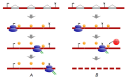
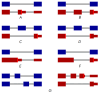
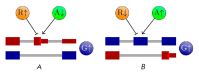
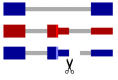
Unproductive splicing is a mechanism of post-transcriptional gene expression control in which premature stop codons are inserted into protein-coding transcripts as a result of regulated alternative splicing, leading to their degradation via the nonsense-mediated decay pathway. This mechanism is especially characteristic of RNA-binding proteins, which regulate each other's expression levels and those of other genes in multiple auto- and cross-regulatory loops. Deregulation of unproductive splicing is a cause of serious human diseases, including cancers, and is increasingly being considered as a prominent therapeutic target. This review discusses the types of unproductive splicing events, the mechanisms of auto- and cross-regulation, nonsense-mediated decay escape, and problems in identifying unproductive splice isoforms. It also provides examples of deregulation of unproductive splicing in human diseases and discusses therapeutic strategies for its correction using antisense oligonucleotides and small molecules.
Keywords: antisense oligonucleotides; nonsense-mediated decay; regulation; splicing; unproductive splicing.
Copyright ® 2024 National Research University Higher School of Economics.
Figures
References
-
- Lykke-Andersen S., Jensen T.H.. Nat. Rev. Mol. Cell Biol. 2015;16(11):665–677. - PubMed
LinkOut - more resources
Full Text Sources
